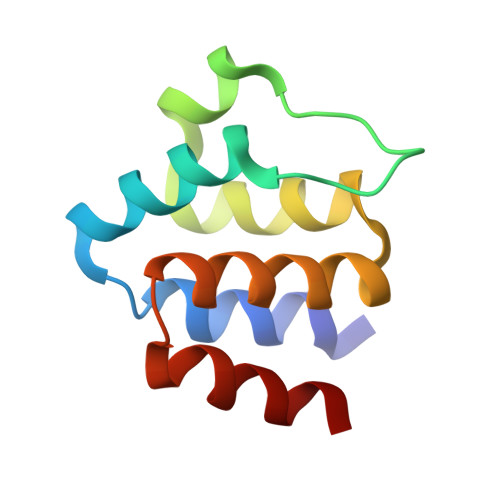
Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Sborgi, L., Ravotti, F., Dandey, V.P., Dick, M.S., Mazur, A., Reckel, S., Chami, M., Scherer, S., Huber, M., Bockmann, A., Egelman, E.H., Stahlberg, H., Broz, P., Meier, B.H., Hiller, S.(2015) Proc Natl Acad Sci U S A 112: 13237-13242
- PubMed: 26464513
- DOI: https://doi.org/10.1073/pnas.1507579112
- Primary Citation of Related Structures:
2N1F - PubMed Abstract:
Inflammasomes are multiprotein complexes that control the innate immune response by activating caspase-1, thus promoting the secretion of cytokines in response to invading pathogens and endogenous triggers. Assembly of inflammasomes is induced by activation of a receptor protein. Many inflammasome receptors require the adapter protein ASC [apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD)], which consists of two domains, the N-terminal pyrin domain (PYD) and the C-terminal CARD. Upon activation, ASC forms large oligomeric filaments, which facilitate procaspase-1 recruitment. Here, we characterize the structure and filament formation of mouse ASC in vitro at atomic resolution. Information from cryo-electron microscopy and solid-state NMR spectroscopy is combined in a single structure calculation to obtain the atomic-resolution structure of the ASC filament. Perturbations of NMR resonances upon filament formation monitor the specific binding interfaces of ASC-PYD association. Importantly, NMR experiments show the rigidity of the PYD forming the core of the filament as well as the high mobility of the CARD relative to this core. The findings are validated by structure-based mutagenesis experiments in cultured macrophages. The 3D structure of the mouse ASC-PYD filament is highly similar to the recently determined human ASC-PYD filament, suggesting evolutionary conservation of ASC-dependent inflammasome mechanisms.
Organizational Affiliation:
Biozentrum, University of Basel, 4056 Basel, Switzerland;