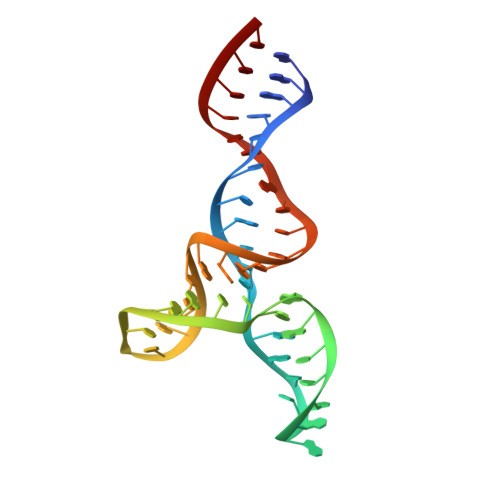
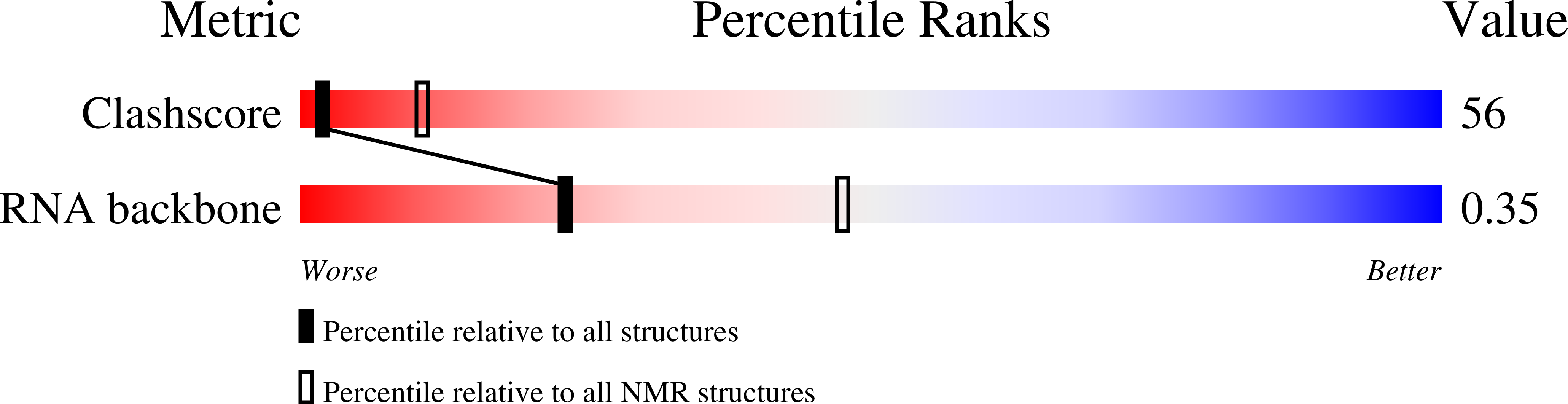The structural stabilization of the kappa three-way junction by Mg(II) represents the first step in the folding of a group II intron.
Donghi, D., Pechlaner, M., Finazzo, C., Knobloch, B., Sigel, R.K.O.(2013) Nucleic Acids Res 41: 2489-2504
- PubMed: 23275550
- DOI: https://doi.org/10.1093/nar/gks1179
- Primary Citation of Related Structures:
2LU0 - PubMed Abstract:
Folding of group II introns is characterized by a first slow compaction of domain 1 (D1) followed by the rapid docking of other domains to this scaffold. D1 compaction initiates in a small subregion encompassing the κ and ζ elements. These two tertiary elements are also the major interaction sites with domain 5 to form the catalytic core. Here, we provide the first characterization of the structure adopted at an early folding step and show that the folding control element can be narrowed down to the three-way junction with the κ motif. In our nuclear magnetic resonance studies of this substructure derived from the yeast mitochondrial group II intron Sc.ai5γ, we show that a high affinity Mg(II) ion stabilizes the κ element and enables coaxial stacking between helices d' and d'', favoring a rigid duplex across the three-way junction. The κ-element folds into a stable GAAA-tetraloop motif and engages in A-minor interactions with helix d'. The addition of cobalt(III)hexammine reveals three distinct binding sites. The Mg(II)-promoted structural rearrangement and rigidification of the D1 core can be identified as the first micro-step of D1 folding.
Organizational Affiliation:
Institute of Inorganic Chemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.