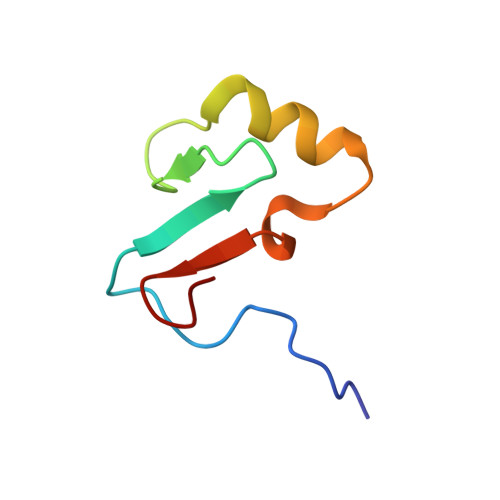
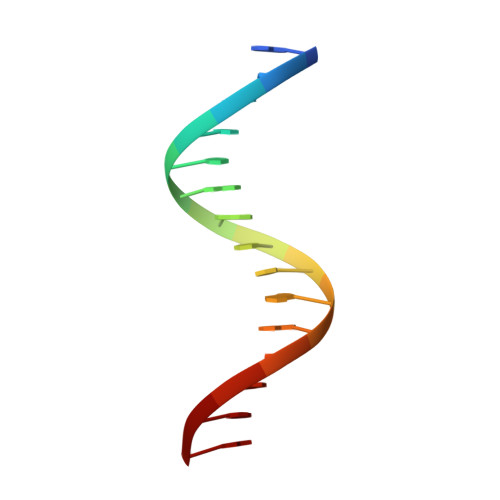
Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Cordeiro, T.N., Schmidt, H., Madrid, C., Juarez, A., Bernado, P., Griesinger, C., Garcia, J., Pons, M.(2011) PLoS Pathog 7: e1002380-e1002380
- PubMed: 22114557
- DOI: https://doi.org/10.1371/journal.ppat.1002380
- Primary Citation of Related Structures:
2LEV - PubMed Abstract:
Ler, a member of the H-NS protein family, is the master regulator of the LEE pathogenicity island in virulent Escherichia coli strains. Here, we determined the structure of a complex between the DNA-binding domain of Ler (CT-Ler) and a 15-mer DNA duplex. CT-Ler recognizes a preexisting structural pattern in the DNA minor groove formed by two consecutive regions which are narrower and wider, respectively, compared with standard B-DNA. The compressed region, associated with an AT-tract, is sensed by the side chain of Arg90, whose mutation abolishes the capacity of Ler to bind DNA. The expanded groove allows the approach of the loop in which Arg90 is located. This is the first report of an experimental structure of a DNA complex that includes a protein belonging to the H-NS family. The indirect readout mechanism not only explains the capacity of H-NS and other H-NS family members to modulate the expression of a large number of genes but also the origin of the specificity displayed by Ler. Our results point to a general mechanism by which horizontally acquired genes may be specifically recognized by members of the H-NS family.
Organizational Affiliation:
Institute for Research in Biomedicine (IRB Barcelona), Parc Científic de Barcelona, Barcelona, Spain.