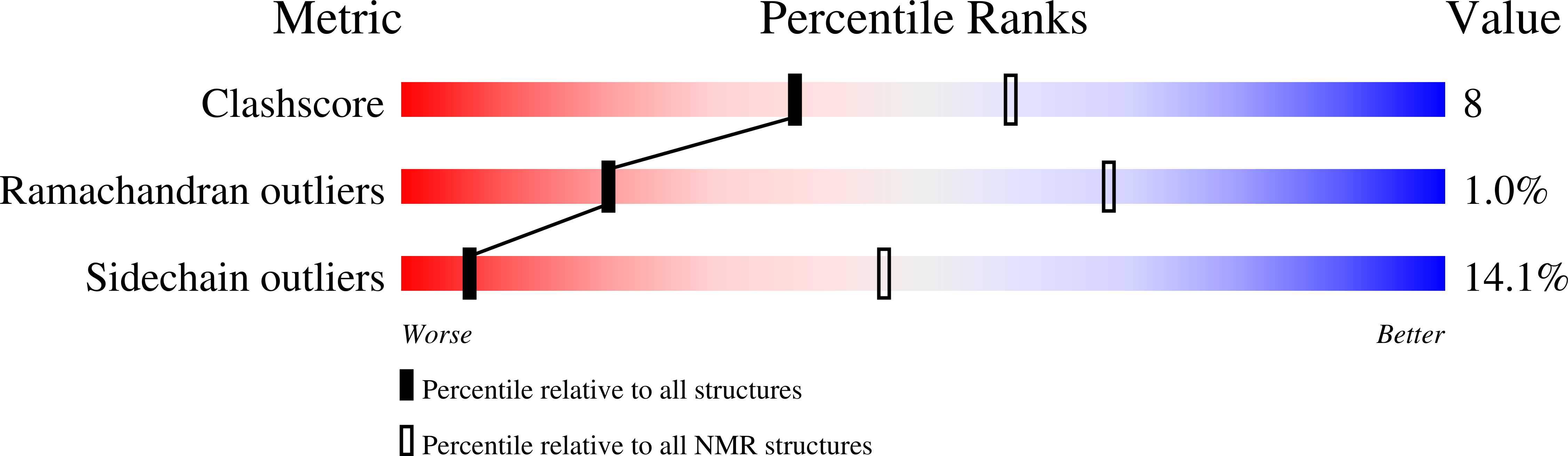Characterization of the PilN, PilO and PilP type IVa pilus subcomplex.
Tammam, S., Sampaleanu, L.M., Koo, J., Sundaram, P., Ayers, M., Andrew Chong, P., Forman-Kay, J.D., Burrows, L.L., Howell, P.L.(2011) Mol Microbiol 82: 1496-1514
- PubMed: 22053789
- DOI: https://doi.org/10.1111/j.1365-2958.2011.07903.x
- Primary Citation of Related Structures:
2LC4 - PubMed Abstract:
Type IVa pili are bacterial nanomachines required for colonization of surfaces, but little is known about the organization of proteins in this system. The Pseudomonas aeruginosa pilMNOPQ operon encodes five key members of the transenvelope complex facilitating pilus function. While PilQ forms the outer membrane secretin pore, the functions of the inner membrane-associated proteins PilM/N/O/P are less well defined. Structural characterization of a stable C-terminal fragment of PilP (PilP(Δ71)) by NMR revealed a modified β-sandwich fold, similar to that of Neisseria meningitidis PilP, although complementation experiments showed that the two proteins are not interchangeable likely due to divergent surface properties. PilP is an inner membrane putative lipoprotein, but mutagenesis of the putative lipobox had no effect on the localization and function of PilP. A larger fragment, PilP(Δ18-6His), co-purified with a PilN(Δ44)/PilO(Δ51) heterodimer as a stable complex that eluted from a size exclusion chromatography column as a single peak with a molecular weight equivalent to two heterotrimers with 1:1:1 stoichiometry. Although PilO forms both homodimers and PilN-PilO heterodimers, PilP(Δ18-6His) did not interact stably with PilO(Δ51) alone. Together these data demonstrate that PilN/PilO/PilP interact directly to form a stable heterotrimeric complex, explaining the dispensability of PilP's lipid anchor for localization and function.
Organizational Affiliation:
Program in Molecular Structure and Function, The Hospital for Sick Children, Toronto, Ontario, M5G 1X8, Canada.