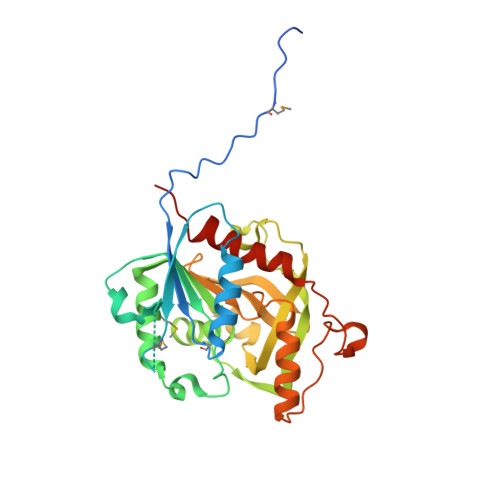
Crystal structure of homoserine O-succinyltransferase from Bacillus cereus at 2.4 A resolution
Zubieta, C., Krishna, S.S., McMullan, D., Miller, M.D., Abdubek, P., Agarwalla, S., Ambing, E., Astakhova, T., Axelrod, H.L., Carlton, D., Chiu, H.J., Clayton, T., Deller, M., Didonato, M., Duan, L., Elsliger, M.A., Grzechnik, S.K., Hale, J., Hampton, E., Han, G.W., Haugen, J., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Koesema, E., Kumar, A., Marciano, D., Morse, A.T., Nigoghossian, E., Oommachen, S., Reyes, R., Rife, C.L., Bedem, H.V., Weekes, D., White, A., Xu, Q., Hodgson, K.O., Wooley, J., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2007) Proteins 68: 999-1005
- PubMed: 17546672
- DOI: https://doi.org/10.1002/prot.21208
- Primary Citation of Related Structures:
2GHR
Organizational Affiliation:
Joint Center for Structural Genomics, Stanford University, Menlo Park, California, USA.