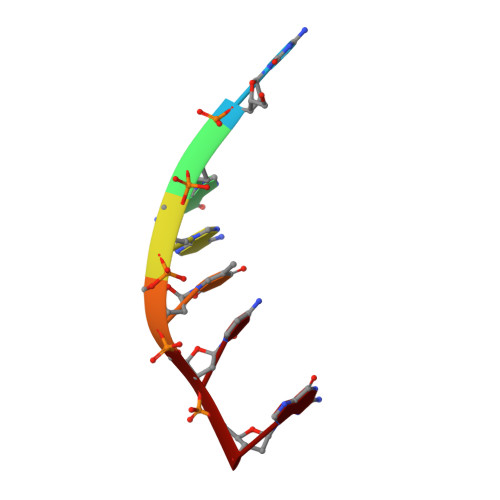
Structure of DNA-porphyrin complex.
Lipscomb, L.A., Zhou, F.X., Presnell, S.R., Woo, R.J., Peek, M.E., Plaskon, R.R., Williams, L.D.(1996) Biochemistry 35: 2818-2823
- PubMed: 8608116
- DOI: https://doi.org/10.1021/bi952443z
- Primary Citation of Related Structures:
231D - PubMed Abstract:
We report the 2.4 A resolution X-ray structure of a complex in which a small molecule flips a base out of a DNA helical stack. The small molecule is a metalloporphyrin, CuTMPyP4 [copper(II) meso-tetra(N-methyl-4-pyridyl)porphyrin], and the DNA is a hexamer duplex, [d(CGATCG)]2. The porphyrin system, with the copper atom near the helical axis, is located within the helical stack. The porphyrin binds by normal intercalation between the C and G of 5' TCG 3' and by extruding the C of 5' CGA 3'. The DNA forms a distorted right-handed helix with only four normal cross-strand Watson-Crick base pairs. Two pyridyl rings are located in each groove of the DNA. The complex appears to be extensively stabilized by electrostatic interactions between positively charged nitrogen atoms of the pyridyl rings and negatively charged phosphate oxygen atoms of the DNA. Favorable electrostatic interactions appear to draw the porphyrin into the duplex interior, offsetting unfavorable steric clashes between the pyridyl rings and the DNA backbone. These pyridyl-backbone clashes extend the DNA along its axis and preclude formation of van der Waals stacking contacts in the interior of the complex. Stacking contacts are the primary contributor to stability of DNA. The unusual lack of van der Waals stacking contacts in the porphyrin complex destabilizes the DNA duplex and decreases the energetic cost of local melting. Thus extrusion of a base appears to be facilitated by pyridyl-DNA steric clashes.
Organizational Affiliation:
School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, 30332-0400, USA.