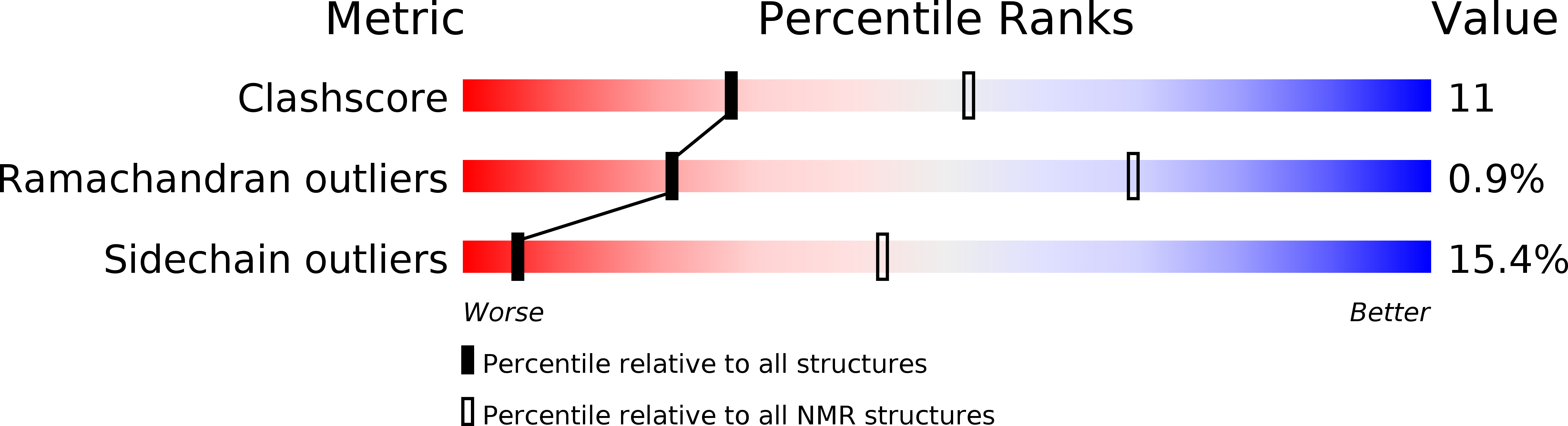Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Barnham, K.J., Monks, S.A., Hinds, M.G., Azad, A.A., Norton, R.S.(1997) Biochemistry 36: 5970-5980
- PubMed: 9166767
- DOI: https://doi.org/10.1021/bi9629945
- Primary Citation of Related Structures:
1ZEC - PubMed Abstract:
Nef is a 27 kDa myristylated phosphoprotein expressed early in infection by HIV. The N terminus of Nef is thought to play a vital role in the functions of this protein through its interactions with membrane structures. The solution structure of a 25-residue polypeptide corresponding to the N terminus of Nef (Nef1-25) has been investigated by 1H NMR spectroscopy. In aqueous solution at pH 4.8 and 281 K, this peptide underwent conformational averaging, with Pro13 existing in cis and trans conformations in nearly equal proportions. In methanol solution, however, the peptide adopted a well-defined alpha-helical structure from residues 6 to 22, with the N- and C-terminal regions having a less ordered structure. On the basis of a comparison of chemical shifts and NOEs, it appeared that this helical structure was maintained in aqueous trifluoroethanol (50% v/v) and to a lesser extent in a solution of SDS micelles. When the N-acetyl group was replaced by either an N-myristyl or a free ammonium group, there was little effect on the three-dimensional structure of the peptide in methanol; deamidation of the C terminus also had no effect on the structure in methanol. In water, the myristylated peptide aggregated. The similarity between the sequences of Nef1-25 and melittin is reflected in the similar structures of the two molecules, although the N-terminal helix of melittin is more defined. This similarity in structure raises the possibility that Nef1-25 not only interacts with membranes but also may be capable of disrupting them and causing cell lysis. This type of interaction could contribute at least in part to the killing of bystander cells in lymphoid tissues during HIV infection.
Organizational Affiliation:
Biomolecular Research Institute, Parkville, Victoria, Australia.