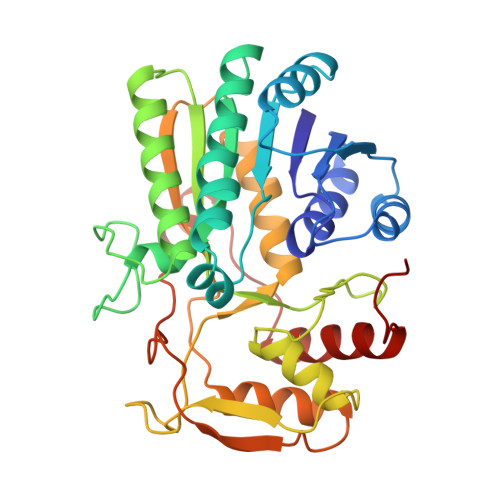
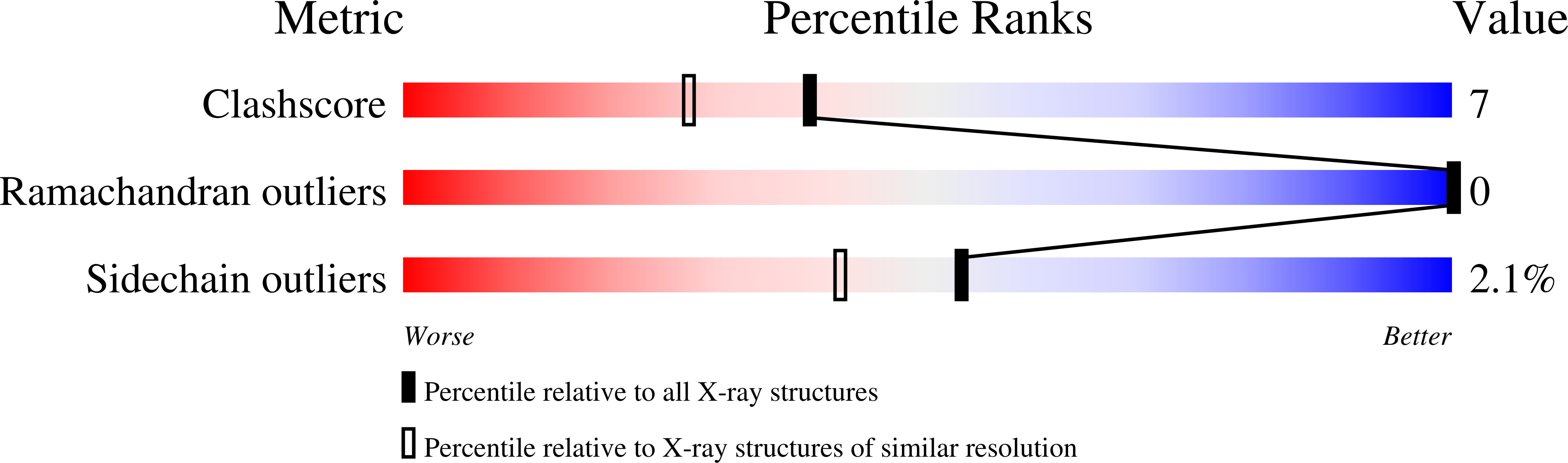Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism.
Thoden, J.B., Frey, P.A., Holden, H.M.(1996) Biochemistry 35: 5137-5144
- PubMed: 8611497
- DOI: https://doi.org/10.1021/bi9601114
- Primary Citation of Related Structures:
1XEL - PubMed Abstract:
UDP-galactose 4-epimerase is one of three enzymes in the metabolic pathway that converts galactose into glucose1-phosphate. Specifically this enzyme catalyzes the interconversion of UDP-galactose and UDP-glucose. The molecular structure of the NADH/UDP-glucose abortive complex of the enzyme from Escherichia coli has been determined by X-ray diffraction analysis to a nominal resolution of 1.8 A and refined to an R-factor of 18.2% for all measurement X-ray data. The nicotinamide ring of the dinucleotide adopts the syn conformation in relationship to the ribose. Both the NADH and UDP-glucose are in the proper orientation for a B-side specific transfer from C4 of the sugar to C4 of the dinucleotide. Those residues implicated in glucose binding include Ser 124, tyr 149, Asn 179, Asn199, Arg 231, and Tyr 299. An amino acid sequence alignment of various prokaryotic and eukaryotic epimerases reveals a high degree of conservation with respect to those residues involved in both NADH and substrate binding. The nonstereospecificity displayed by epimerase was originally thought to occur through a simple rotation about the bond between the glycosyl C1 oxygen of the 4-ketose intermediate and the beta-phosphorous of the UDP moiety, thereby allowing the opposite side of the sugar to face the NADH. The present structure reveals that additional rotations about the phosphate backbone of UDP are necessary. Furthermore, the abortive complex model described here suggests that Ser 124 and Tyr 149 are likely to play important roles in the catalytic mechanism of the enzyme.
Organizational Affiliation:
Institute for Enzyme Research, Graduate School, University of Wisconsin, Madison 53705, USA.