Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Guarne, A., Ramon-Maiques, S., Wolff, E.M., Ghirlando, R., Hu, X., Miller, J.H., Yang, W.(2004) EMBO J 23: 4134-4145
- PubMed: 15470502
- DOI: https://doi.org/10.1038/sj.emboj.7600412
- Primary Citation of Related Structures:
1X9Z - PubMed Abstract:
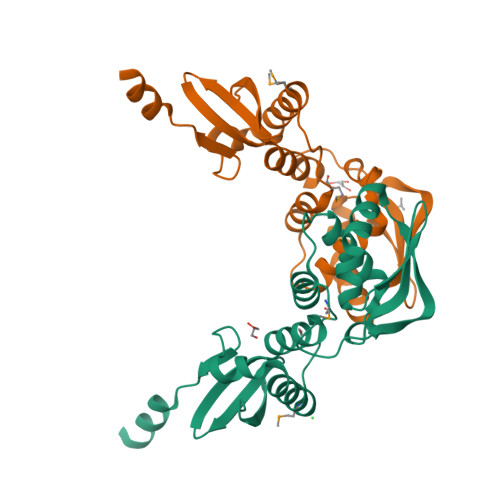
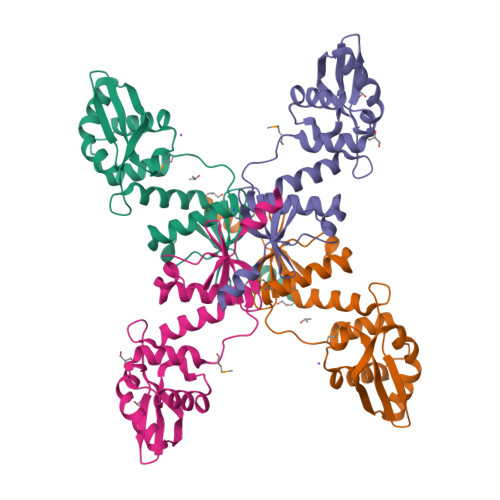
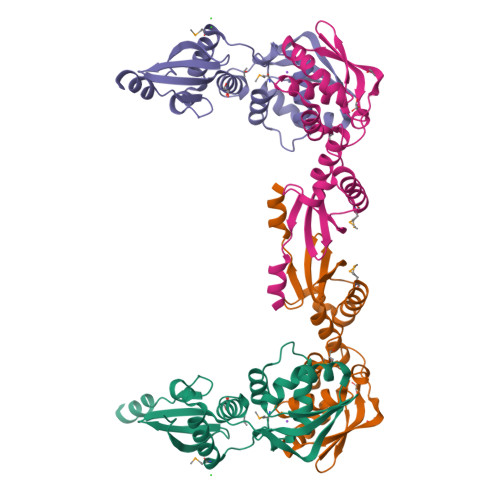
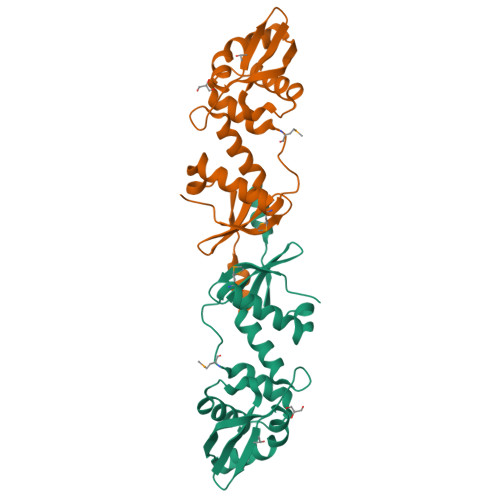
MutL assists the mismatch recognition protein MutS to initiate and coordinate mismatch repair in species ranging from bacteria to humans. The MutL N-terminal ATPase domain is highly conserved, but the C-terminal region shares little sequence similarity among MutL homologs. We report here the crystal structure of the Escherichia coli MutL C-terminal dimerization domain and the likelihood of its conservation among MutL homologs. A 100-residue proline-rich linker between the ATPase and dimerization domains, which generates a large central cavity in MutL dimers, tolerates sequence substitutions and deletions of one-third of its length with no functional consequences in vivo or in vitro. Along the surface of the central cavity, residues essential for DNA binding are located in both the N- and C-terminal domains. Each domain of MutL interacts with UvrD helicase and is required for activating the helicase activity. The DNA-binding capacity of MutL is correlated with the level of UvrD activation. A model of how MutL utilizes its ATPase and DNA-binding activities to mediate mismatch-dependent activation of MutH endonuclease and UvrD helicase is proposed.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.