How the Human Telomeric Proteins Trf1 and Trf2 Recognize Telomeric DNA: A View from High-Resolution Crystal Structures
Court, R.I., Chapman, L.M., Fairall, L., Rhodes, D.(2005) EMBO Rep 6: 39
- PubMed: 15608617
- DOI: https://doi.org/10.1038/sj.embor.7400314
- Primary Citation of Related Structures:
1W0T, 1W0U - PubMed Abstract:
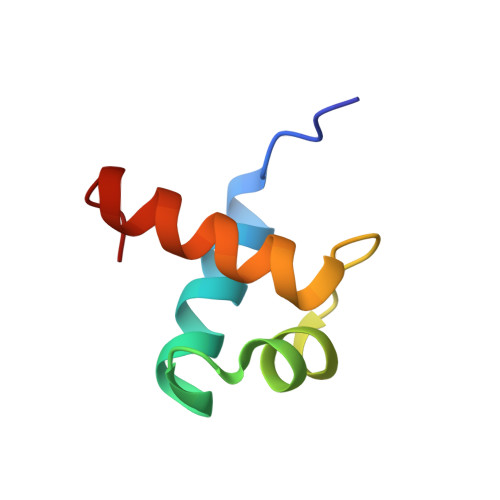
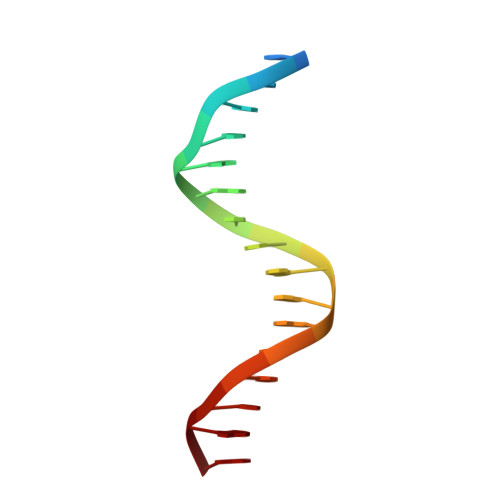
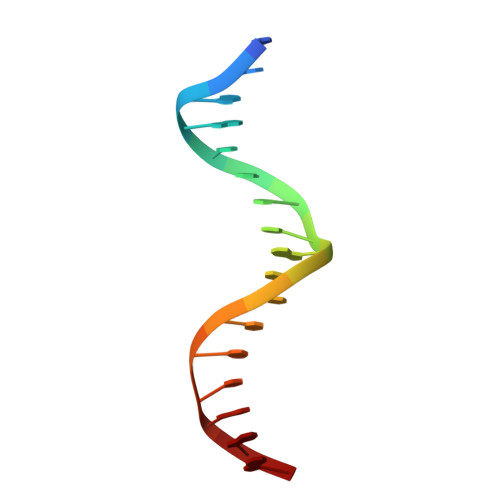
Human telomeres consist of tandem arrays of TTAGGG sequence repeats that are specifically bound by two proteins, TRF1 and TRF2. They bind to DNA as preformed homodimers and have the same architecture in which the DNA-binding domains (Dbds) form independent structural units. Despite these similarities, TRF1 and TRF2 have different functions at telomeres. The X-ray crystal structures of both TRF1- and TRF2-Dbds in complex with telomeric DNA (2.0 and 1.8 angstroms resolution, respectively) show that they recognize the same TAGGGTT binding site by means of homeodomains, as does the yeast telomeric protein Rap1p. Two of the three G-C base pairs that characterize telomeric repeats are recognized specifically and an unusually large number of water molecules mediate protein-DNA interactions. The binding of the TRF2-Dbd to the DNA double helix shows no distortions that would account for the promotion of t-loops in which TRF2 has been implicated.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.