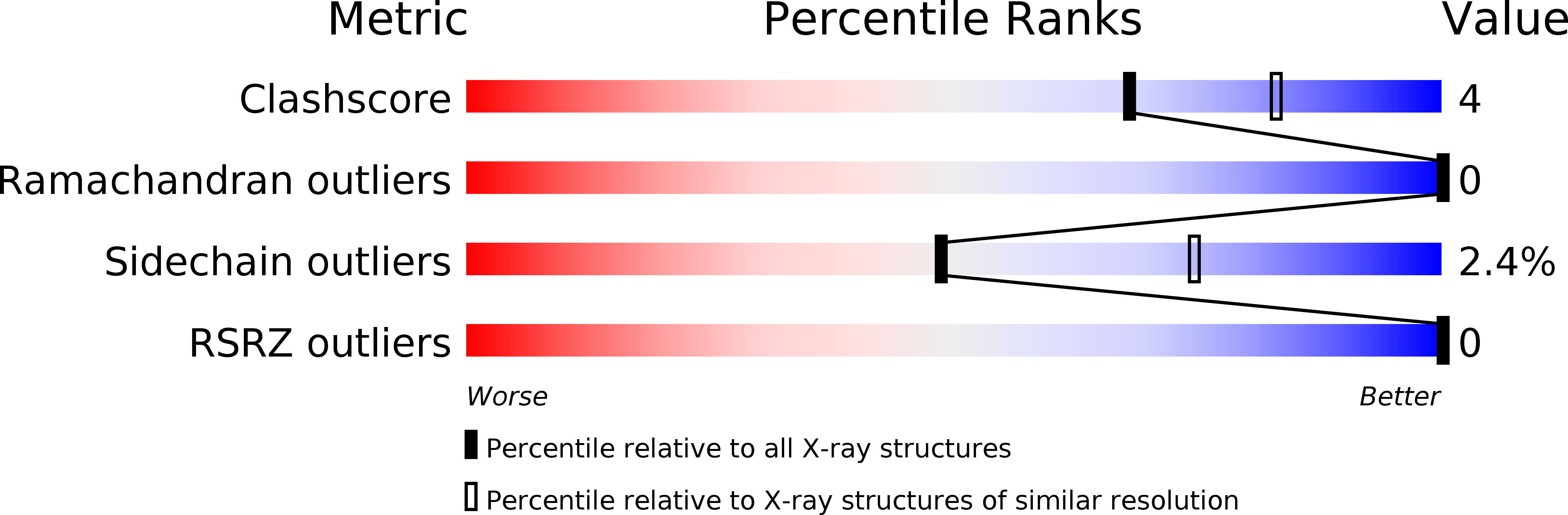
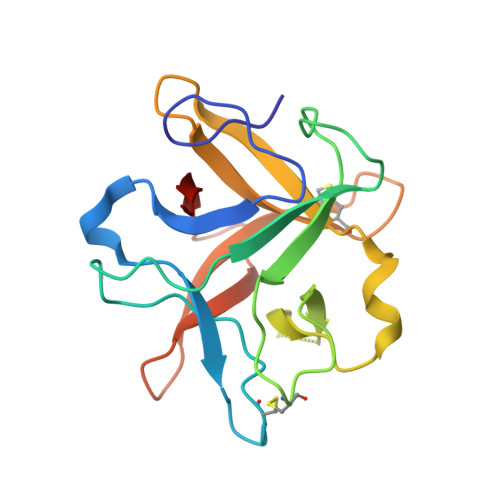
Crystal structure of a Kunitz-type trypsin inhibitor from Erythrina caffra seeds.
Onesti, S., Brick, P., Blow, D.M.(1991) J Mol Biol 217: 153-176
- PubMed: 1988676
- DOI: https://doi.org/10.1016/0022-2836(91)90618-g
- Primary Citation of Related Structures:
1TIE - PubMed Abstract:
The trypsin inhibitor DE-3 from Erythrina caffra (ETI) belongs to the Kunitz-type soybean trypsin inhibitor (STI) family and consists of 172 amino acid residues with two disulphide bridges. The amino acid sequence of ETI shows high homology to other trypsin inhibitors from the same family but ETI has the unique ability to bind and inhibit tissue plasminogen activator. The crystal structure of ETI has been determined using the method of isomorphous replacement and refined using a combination of simulated annealing and conventional restrained least-squares crystallographic refinement. The refined model includes 60 water molecules and 166 amino acid residues, with a root-mean-square deviation in bond lengths from ideal values of 0.016 A. The crystallographic R-factor is 20.8% for 7770 independent reflections between 10.0 and 2.5 A. The three-dimensional structure of ETI consists of 12 antiparallel beta-strands joined by long loops. Six of the strands form a short antiparallel beta-barrel that is closed at one end by a "lid" consisting of the other six strands coupled in pairs. The molecule shows approximate 3-fold symmetry about the axis of the barrel, with the repeating unit consisting of four sequential beta-strands and the connecting loops. Although there is no sequence homology, this same fold is present in the structure of interleukin-1 alpha and interleukin-1 beta. When the structure of ETI and interleukin-1 beta are superposed, the close agreement between the alpha-carbon positions for the beta-strands is striking. The scissile bond (Arg63-Ser64) is located on an external loop that protrudes from the surface of the molecule and whose architecture is not constrained by secondary structure elements, disulphide bridges or strong electrostatic interactions. The hydrogen bonds made by the side-chain amide group of Asn12 play a key role in maintaining the three-dimensional structure of the loop. This residue is in a position corresponding to that of a conserved asparagine in the Kazal inhibitor family. Although the overall structure of ETI is similar to the partial structure of STI, the scissile bond loop is displaced by about 4 A. This displacement probably arises from the fact that the structure of STI has been determined in a complex with trypsin but could possibly be a consequence of the close molecular contact between Arg63 and an adjacent molecule in the crystal lattice.
Organizational Affiliation:
Blackett Laboratory, Imperial College, London, England, U.K.