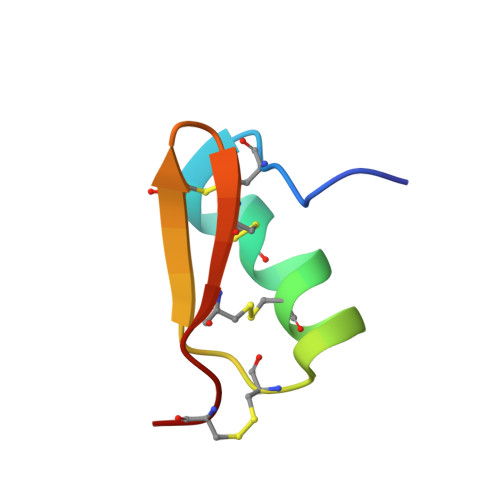
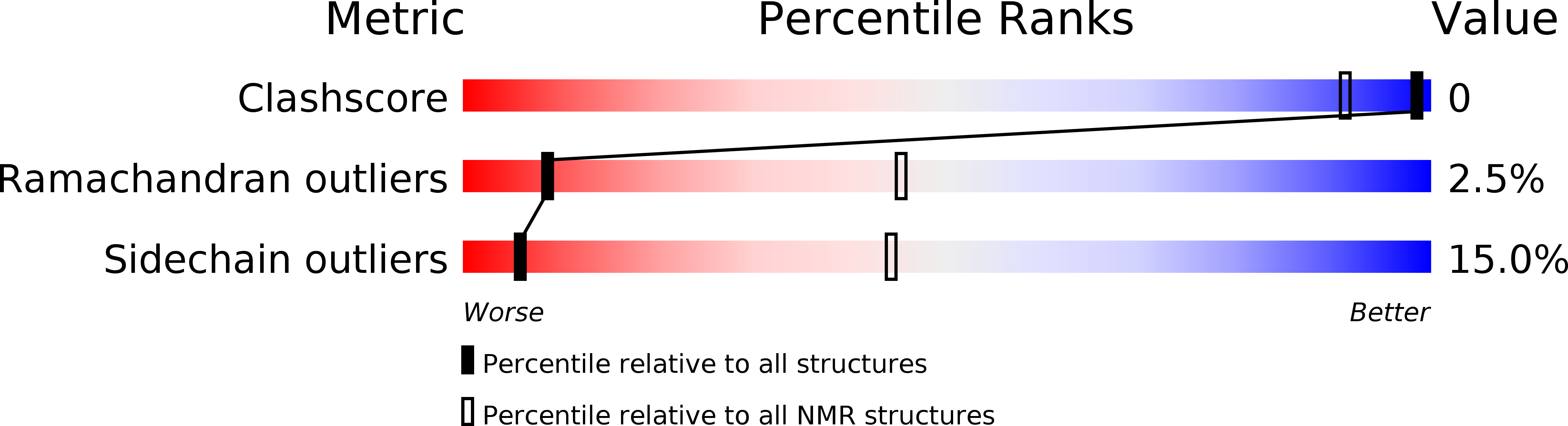Pi7, an orphan peptide from the scorpion Pandinus imperator: a 1H-NMR analysis using a nano-NMR Probe.
Delepierre, M., Prochnicka-Chalufour, A., Boisbouvier, J., Possani, L.D.(1999) Biochemistry 38: 16756-16765
- PubMed: 10606507
- DOI: https://doi.org/10.1021/bi991685m
- Primary Citation of Related Structures:
1QKY - PubMed Abstract:
The three-dimensional solution structure of a novel peptide, Pi7, purified from the venom of the scorpion Pandinus imperator, and for which no specific receptor has been found yet, was determined by two-dimensional homonuclear proton NMR methods from a nanomole amount of compound using a nano-nmr probe. Pandinus imperator peptide 7 does not block voltage-dependent K(+)-channels and does not displace labeled noxiustoxin from rat brain synaptosomal membranes. The toxin has 38 amino acid residues and, similarly to Pi1, is stabilized by four disulfide bridges (Cys6-Cys27, Cys12-Cys32, Cys16-Cys34, and Cys22-Cys37). In addition, the lysine at position 26 crucial for potassium-channel blocking is replaced in Pi7 by an arginine. Tyrosine 34, equivalent to Tyr36 of ChTX is present, but the N-terminal positions 1 and 2 are occupied by two acidic residues Asp and Glu, respectively. The dihedral angles and distance restraints obtained from measured NMR parameters were used in structural calculations in order to determine the conformation of the peptide. The disulfide-bridge topology was established using distance restraints allowing ambiguous partners between S atoms combined with NMR-derived structural information. The structure is organized around a short alpha-helix spanning residues Thr9 to Thr20/Gly21 and a beta-sheet. These two elements of secondary structure are stabilized by two disulfide bridges, Cys12-Cys32 and Cys16-Cys34. The antiparallel beta-sheet is composed of two strands extending from Asn22 to Cys34 with a tight turn at Ile28-Asn29 in contact with the N-terminal fragment Ile4 to Cys6.
Organizational Affiliation:
Laboratoire de RMN Institut Pasteur, CNRS URA 1129, PARIS, France. murield@pasteur.fr