Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
Egli, M., Tereshko, V., Mushudov, G.N., Sanishvili, R., Liu, X., Lewis, F.D.(2003) J Am Chem Soc 125: 10842-10849
- PubMed: 12952463
- DOI: https://doi.org/10.1021/ja0355527
- Primary Citation of Related Structures:
1PUY - PubMed Abstract:
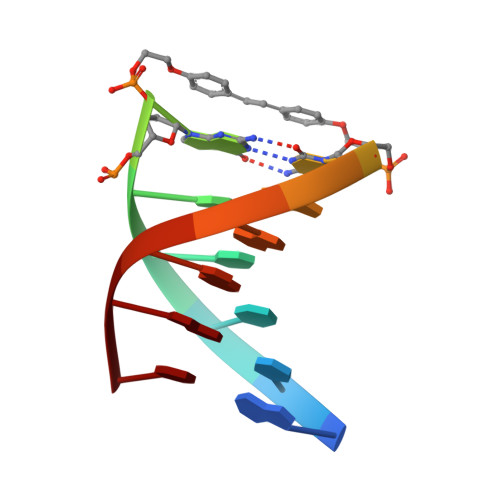
Synthetic conjugates possessing bis(2-hydroxyethyl)stilbene-4,4'-diether linkers (Sd2) form the most stable DNA hairpins reported to date. Factors that affect stability are length and flexibility of the linkers and pi-stacking of the stilbene moiety on the adjacent base pair. The crystal structure of the hairpin d(GT(4)G)-Sd2-d(CA(4)C) was determined at 1.5 A resolution. The conformations of the two molecules in the asymmetric unit differ both in the linker and the stem portions. One of them shows a planar stilbene that is stacked on the adjacent G:C base pair. The other displays considerable rotation between the phenyl rings and an unprecedented edge-to-face orientation of stilbene and base pair. The observation of considerable variations in the conformation of the Sd moiety in the crystal structure allows us to exclude restriction of motion as the reason for the absence of Sd photoisomerization in the hairpins. Conformational differences in the stem portion of the two hairpin molecules go along with different Mg(2+) binding modes. Most remarkable among them is the sequence-specific coordination of a metal ion in the narrow A-tract minor groove. The crystal structure provides unequivocal evidence that a fully hydrated Mg(2+) ion can penetrate the narrow A-tract minor groove, causing the groove to further contract. Overall, the structural data provide a better understanding of the origins of hairpin stability and their photochemical behavior in solution.
Organizational Affiliation:
Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee 37235, USA. martin.egli@vanderbilt.edu