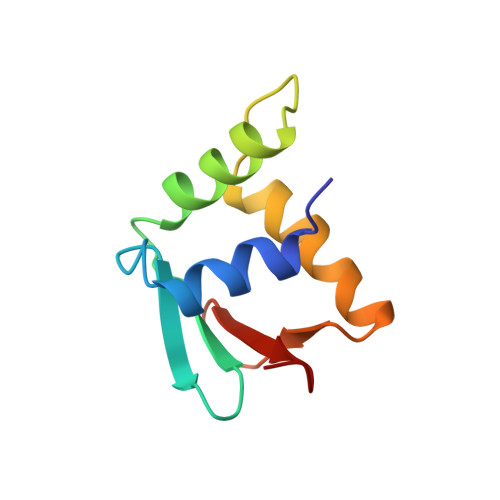
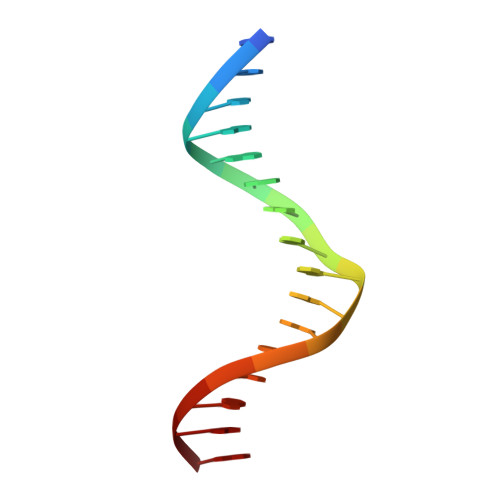
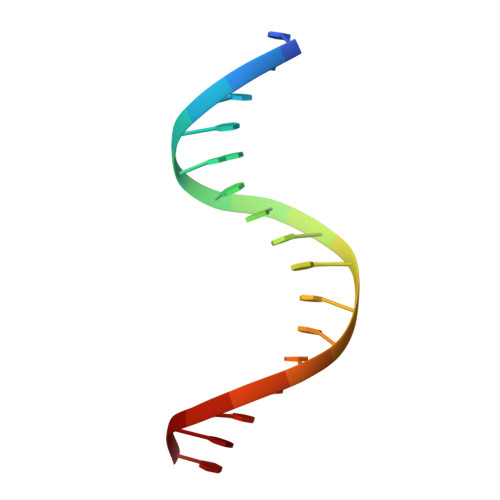
A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Kodandapani, R., Pio, F., Ni, C.Z., Piccialli, G., Klemsz, M., McKercher, S., Maki, R.A., Ely, K.R.(1996) Nature 380: 456-460
- PubMed: 8602247
- DOI: https://doi.org/10.1038/380456a0
- Primary Citation of Related Structures:
1PUE - PubMed Abstract:
The Ets family of transcription factors, of which there are now about 35 members regulate gene expression during growth and development. They share a conserved domain of around 85 amino acids which binds as a monomer to the DNA sequence 5'-C/AGGAA/T-3'. We have determined the crystal structure of an ETS domain complexed with DNA, at 2.3-A resolution. The domain is similar to alpha + beta (winged) 'helix-turn-helix' proteins and interacts with a ten-base-pair region of duplex DNA which takes up a uniform curve of 8 degrees. The domain contacts the DNA by a novel loop-helix-loop architecture. Four of amino acids that directly interact with the DNA are highly conserved: two arginines from the recognition helix lying in the major groove, one lysine from the 'wing' that binds upstream of the core GGAA sequence, and another lysine, from the 'turn' of the 'helix-turn-helix' motif, which binds downstream and on the opposite strand.
Organizational Affiliation:
La Jolla Cancer Research Center at the Burnham Institute, California 92037, USA.