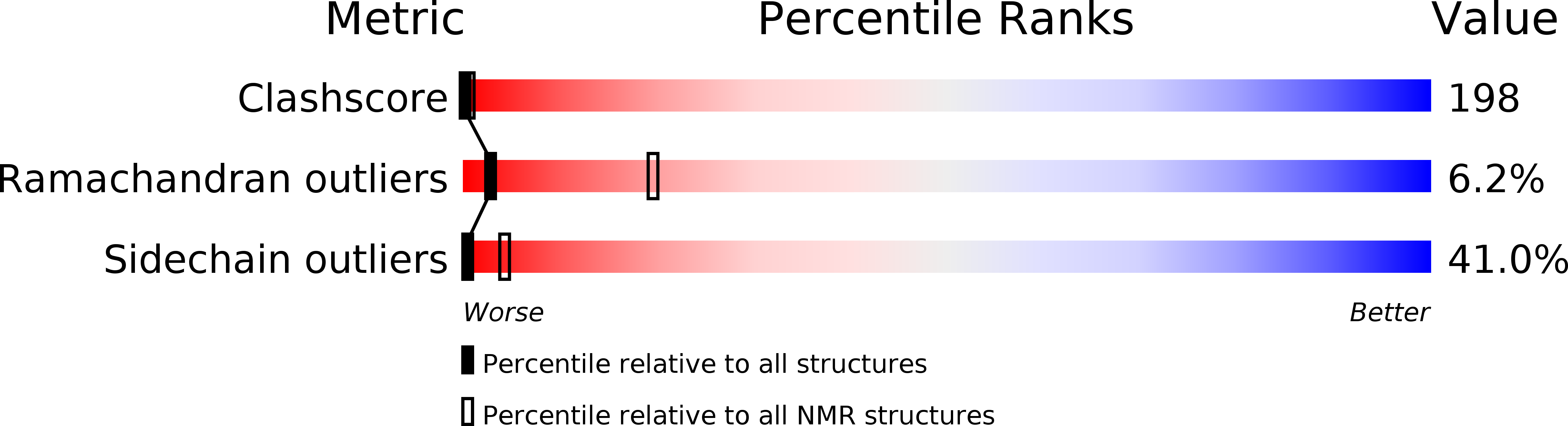A refined model for the solution structure of oxidized putidaredoxin.
Pochapsky, T.C., Jain, N.U., Kuti, M., Lyons, T.A., Heymont, J.(1999) Biochemistry 38: 4681-4690
- PubMed: 10200155
- DOI: https://doi.org/10.1021/bi983030b
- Primary Citation of Related Structures:
1PDX - PubMed Abstract:
A refined model for the solution structure of oxidized putidaredoxin (Pdxo), a Cys4Fe2S2 ferredoxin, has been determined. A previous structure (Pochapsky et al. (1994) Biochemistry 33, 6424-6432; PDB entry ) was calculated using the results of homonuclear two-dimensional NMR experiments. New data has made it possible to calculate a refinement of the original Pdxo solution structure. First, essentially complete assignments for diamagnetic 15N and 13C resonances of Pdxo have been made using multidimensional NMR methods, and 15N- and 13C-resolved NOESY experiments have permitted the identification of many new NOE restraints for structural calculations. Stereospecific assignments for leucine and valine CH3 resonances were made using biosynthetically directed fractional 13C labeling, improving the precision of NOE restraints involving these residues. Backbone dihedral angle restraints have been obtained using a combination of two-dimensional J-modulated 15N,1H HSQC and 3D (HN)CO(CO)NH experiments. Second, the solution structure of a diamagnetic form of Pdx, that of the C85S variant of gallium putidaredoxin, in which a nonligand Cys is replaced by Ser, has been determined (Pochapsky et al. (1998) J. Biomol. NMR 12, 407-415), providing information concerning structural features not observable in the native ferredoxin due to paramagnetism. Third, a crystal structure of a closely related ferredoxin, bovine adrenodoxin, has been published (Müller et al. (1998) Structure 6, 269-280). This structure has been used to model the metal binding site structure in Pdx. A family of fourteen structures is presented that exhibits an rmsd of 0.51 A for backbone heavy atoms and 0.83 A for all heavy atoms. Exclusion of the modeled metal binding loop region reduces overall the rmsd to 0.30 A for backbone atoms and 0.71 A for all heavy atoms.
Organizational Affiliation:
Departments of Chemistry and Biology, Brandeis University, Waltham, Massachusetts 02254-9110, USA. pochapsky@binah.cc.brandeis.edu