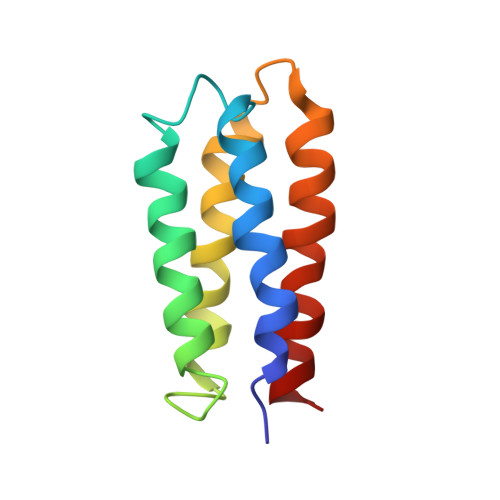
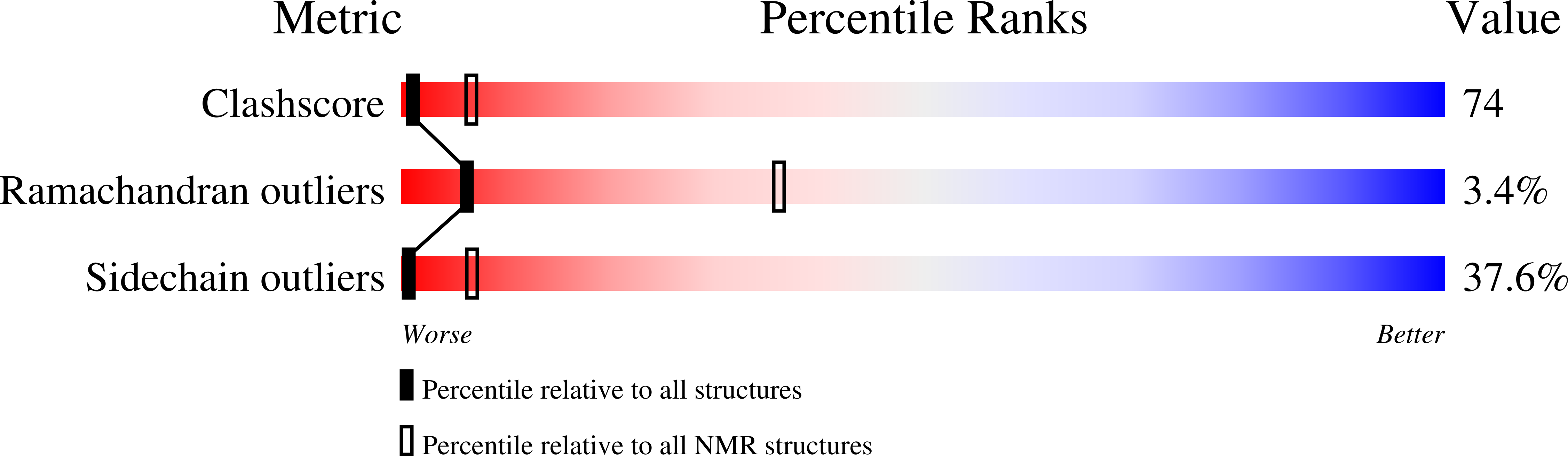Solution structure of a de novo protein from a designed combinatorial library.
Wei, Y., Kim, S., Fela, D., Baum, J., Hecht, M.H.(2003) Proc Natl Acad Sci U S A 100: 13270-13273
- PubMed: 14593201
- DOI: https://doi.org/10.1073/pnas.1835644100
- Primary Citation of Related Structures:
1P68 - PubMed Abstract:
Combinatorial libraries of de novo amino acid sequences can provide a rich source of diversity for the discovery of novel proteins. Randomly generated sequences, however, rarely fold into well ordered protein-like structures. To enhance the quality of a library, diversity must be focused into those regions of sequence space most likely to yield well folded structures. We have constructed focused libraries of de novo sequences by designing the binary pattern of polar and nonpolar amino acids to favor structures that contain abundant secondary structure, while simultaneously burying hydrophobic side chains in the protein interior and exposing hydrophilic side chains to solvent. Because binary patterning specifies only the polar/nonpolar periodicity, but not the identities of the side chains, detailed structural features, including packing interactions, cannot be designed a priori. Can binary patterned libraries nonetheless encode well folded proteins? An unambiguous answer to this question requires determination of a 3D structure. We used NMR spectroscopy to determine the structure of S-824, a novel protein from a recently constructed library of 102-residue sequences. This library is "naïve" in that it has not been subjected to high-throughput screens or directed evolution. The experimentally determined structure of S-824 is a four-helix bundle, as specified by the design. As dictated by the binary-code strategy, nonpolar side chains are buried in the protein interior, and polar side chains are exposed to solvent. The polypeptide backbone and buried side chains are well ordered, demonstrating that S-824 is not a molten globule and forms a unique structure. These results show that amino acid sequences that have neither been selected by evolution, nor designed by computer, nor isolated by high-throughput screening, can form native-like structures. These findings validate the binary-code strategy as an effective method for producing vast collections of well folded de novo proteins.
Organizational Affiliation:
Department of Chemistry, Princeton University, Princeton, NJ 08544, USA.