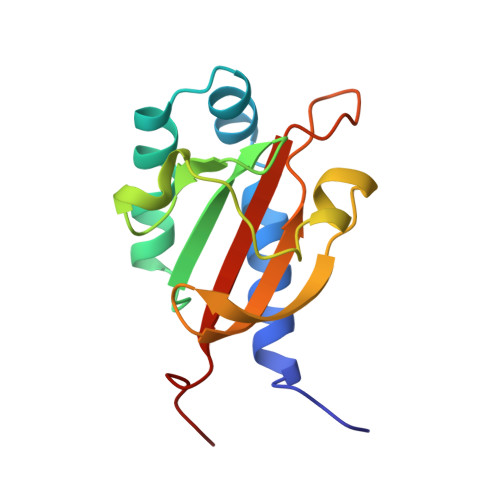
The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
Reinelt, S., Hofmann, E., Gerharz, T., Bott, M., Madden, D.R.(2003) J Biol Chem 278: 39189-39196
- PubMed: 12867417
- DOI: https://doi.org/10.1074/jbc.M305864200
- Primary Citation of Related Structures:
1P0Z - PubMed Abstract:
The integral membrane sensor kinase CitA of Klebsiella pneumoniae is part of a two-component signal transduction system that regulates the transport and metabolism of citrate in response to its environmental concentration. Two-component systems are widely used by bacteria for such adaptive processes, but the stereochemistry of periplasmic ligand binding and the mechanism of signal transduction across the membrane remain poorly understood. The crystal structure of the CitAP periplasmic sensor domain in complex with citrate reveals a PAS fold, a versatile ligand-binding structural motif that has not previously been observed outside the cytoplasm or implicated in the transduction of conformational signals across the membrane. Citrate is bound in a pocket that is shared among many PAS domains but that shows structural variation according to the nature of the bound ligand. In CitAP, some of the citrate contact residues are located in the final strand of the central beta-sheet, which is connected to the C-terminal transmembrane helix. These secondary structure elements thus provide a potential conformational link between the periplasmic ligand binding site and the cytoplasmic signaling domains of the receptor.
Organizational Affiliation:
Ion Channel Structure Group, Max Planck Institute for Medical Research, D-69120 Heidelberg, Germany.