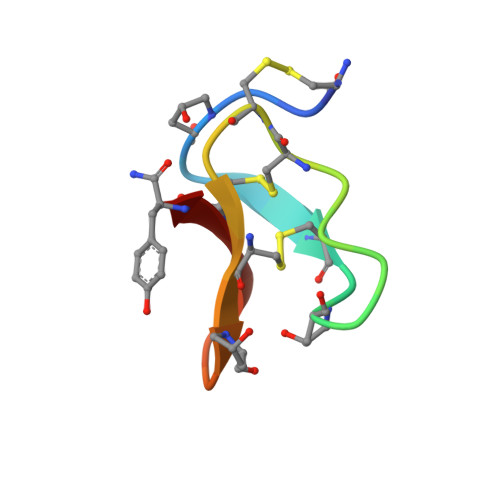
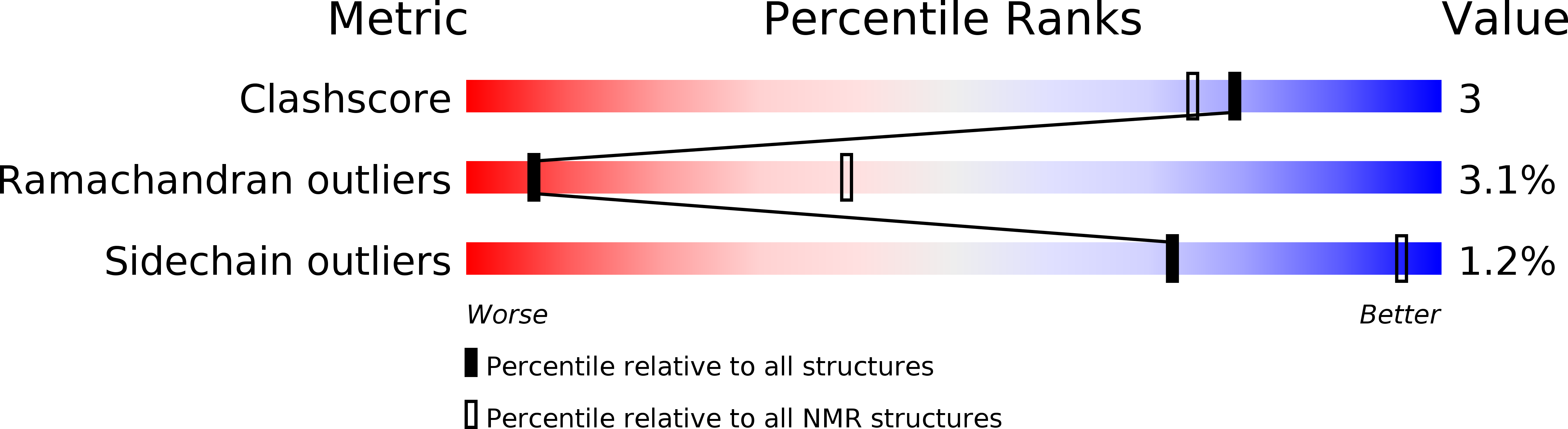Solution structure of omega-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis.
Davis, J.H., Bradley, E.K., Miljanich, G.P., Nadasdi, L., Ramachandran, J., Basus, V.J.(1993) Biochemistry 32: 7396-7405
- PubMed: 8338837
- DOI: https://doi.org/10.1021/bi00080a009
- Primary Citation of Related Structures:
1OMC - PubMed Abstract:
We report here the solution structure of omega-conotoxin GVIA, a peptide antagonist of the N-type neuronal voltage-sensitive calcium channel. The structure was determined using two-dimensional NMR in combination with distance geometry and restrained molecular dynamics. The full relaxation matrix analysis program MARDIGRAS was used to generate maximum and minimum distance restraints from the crosspeak intensities in NOESY spectra. The 187 restraints obtained were used in conjunction with 23 angle restraints from vicinal coupling constants as input for the structure calculations. The backbones of the best 21 structures match with an average pairwise RMSD of 0.58 A. The structures contain a short segment of triple-stranded beta-sheet involving residues 6-8, 18-21, and 24-27, making this the smallest published peptide structure to contain a triple-stranded beta-sheet. Conotoxins have been shown to be effective neuroprotective agents in animal models of brain ischemia. Our results should aid in the design of novel nonpeptide compounds with potential therapeutic utility.
Organizational Affiliation:
Graduate Group in Biophysics, University of California, San Francisco 94143-0446.