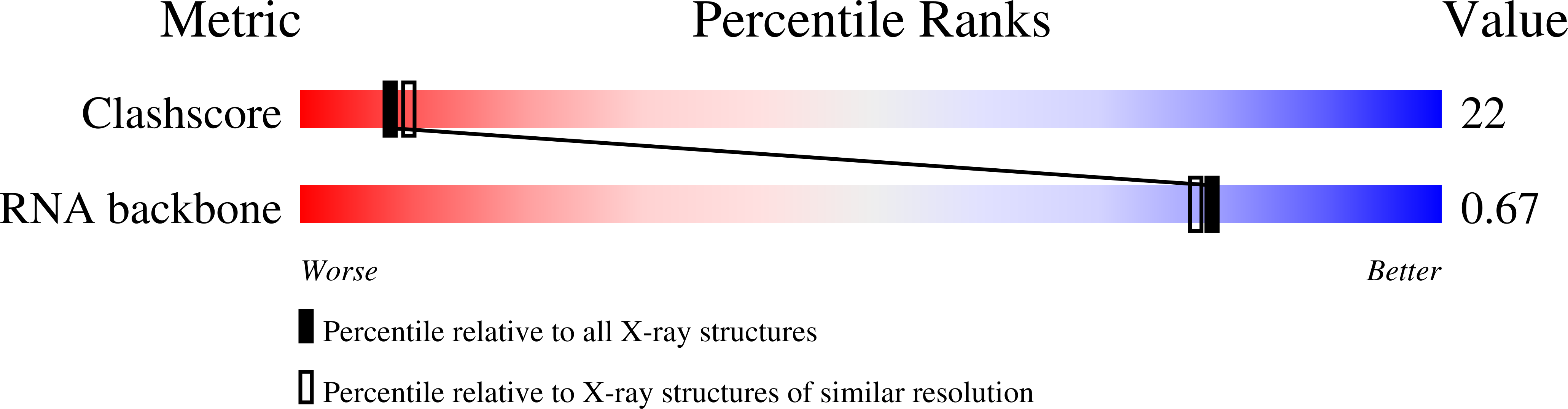The Complex of a Designer Antibiotic with a Model Aminoacyl Site of the 30S Ribosomal Subunit Revealed by X-Ray Crystallography
Russell, R., Murray, J.B., Lentzen, G., Haddad, J., Mobashery, S.(2003) J Am Chem Soc 125: 3410
- PubMed: 12643685
- DOI: https://doi.org/10.1021/ja029736h
- Primary Citation of Related Structures:
1O9M - PubMed Abstract:
The ribosome is an important target for aminoglycoside antibiotics; however, the clinical effectiveness of aminoglycosides has diminished due to bacterial resistance mechanisms. Here we report the X-ray structure of a novel synthetic aminoglycoside bound to the A site of the ribosome, its target for manifestation of activity. The structure validates the in silico design paradigms for the antibiotic and reveals the molecular interactions made by this novel antibiotic in prokaryotes.
Organizational Affiliation:
RiboTargets, Granta Park, Cambridge CB1 6GB, United Kingdom.