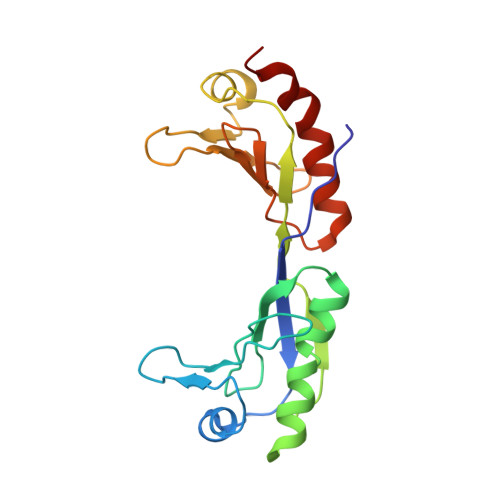
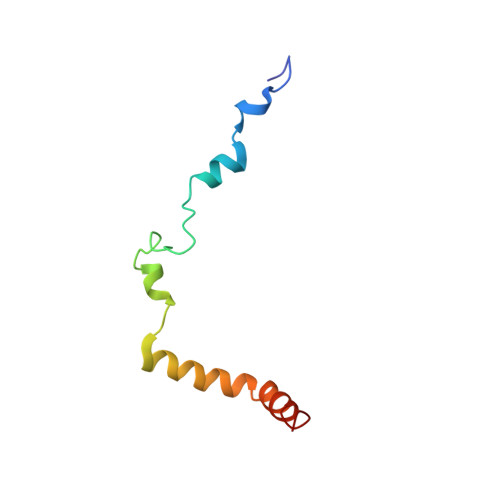
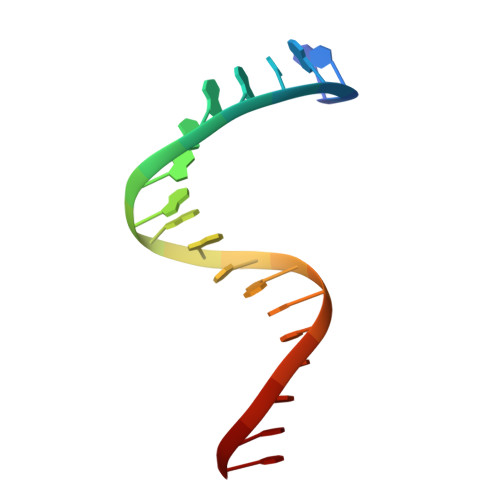
Crystal structure of a transcription factor IIIB core interface ternary complex
Juo, Z.S., Kassavetis, G.A., Wang, J., Geiduschek, E.P., Sigler, P.B.(2003) Nature 422: 534-539
- PubMed: 12660736
- DOI: https://doi.org/10.1038/nature01534
- Primary Citation of Related Structures:
1NGM - PubMed Abstract:
Transcription factor IIIB (TFIIIB), consisting of the TATA-binding protein (TBP), TFIIB-related factor (Brf1) and Bdp1, is a central component in basal and regulated transcription by RNA polymerase III. TFIIIB recruits its polymerase to the promoter and subsequently has an essential role in the formation of the open initiation complex. The amino-terminal half of Brf1 shares a high degree of sequence similarity with the polymerase II general transcription factor TFIIB, but it is the carboxy-terminal half of Brf1 that contributes most of its binding affinity with TBP. The principal anchoring region is located between residues 435 and 545 of yeast Brf1, comprising its homology domain II. The same region also provides the primary interface for assembling Bdp1 into the TFIIIB complex. We report here a 2.95 A resolution crystal structure of the ternary complex containing Brf1 homology domain II, the conserved region of TBP and 19 base pairs of U6 promoter DNA. The structure reveals the core interface for assembly of TFIIIB and demonstrates how the loosely packed Brf1 domain achieves remarkable binding specificity with the convex and lateral surfaces of TBP.
Organizational Affiliation:
Department of Molecular Biophysics & Biochemistry, Yale University, 266 Whitney Avenue, New Haven, Connecticut 06520-8114, USA. juo@csb.yale.edu