Solution Structure of a DNA Duplex Containing Mispair-Aligned N4C-Ethyl-N4C Interstrand Cross-Linked Cytosines
Webba da Silva, M., Noronha, A.M., Noll, D.M., Miller, P.S., Colvin, O.M., Gamcsik, M.P.(2002) Biochemistry 41: 15181-15188
- PubMed: 12484755
- DOI: https://doi.org/10.1021/bi026368l
- Primary Citation of Related Structures:
1N4B - PubMed Abstract:
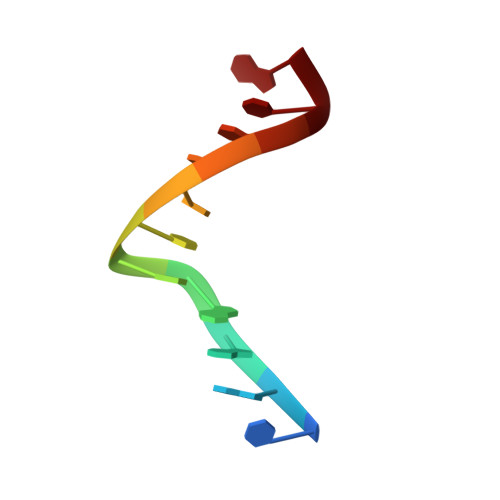
The solution structure of an interstrand cross-linked self-complementary oligodeoxynucleotide containing directly opposed alkylated N(4)C-ethyl-N(4)C cytosine bases was determined by molecular dynamics calculations guided by NMR-derived restraints. The undecamer d(CGAAACTTTCG)(2), where C represents directly opposed alkylated N(4)C-ethyl-N(4)C cytosine bases, serves as model for the cytotoxic cross-links formed by bifunctional alkylating agents used in cancer therapy. The structure of the duplex shows the cross-link protruding into the major groove. An increase in the diameter of the DNA at the pseudoplatform formed by the cross-linked residues creates an A-DNA characteristic hole in the central portion of the DNA. This results in a centrally underwound base step and a number of subsequent overwinding steps leading to an overall axis bend toward the major groove. The structure shows narrowing of both minor and major grooves in the proximity of the cross-link. The perturbation leads to preferential intrastrand base stacking, disruption of adjacent canonical (A.T) base pairing, and buckling of base pairs, the extent of which diminishes with progression away from the lesion site. Overall, the distortion induced by the cross-link spreads over three base pairs on the 5'- and 3'-sides of the cross-link.
Organizational Affiliation:
Department of Medicine, Duke University Medical Center, Durham, North Carolina 27710, USA.