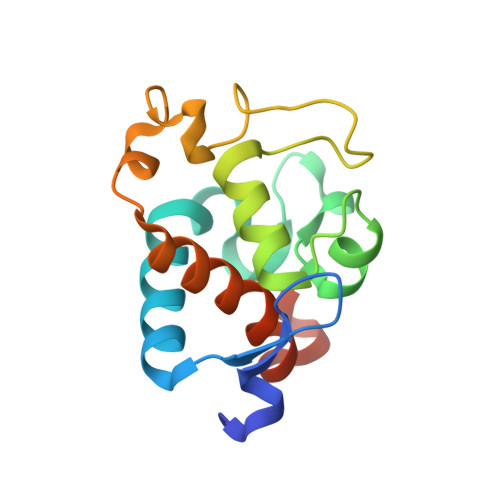
Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Kerfeld, C.A., Sawaya, M.R., Bottin, H., Tran, K.T., Sugiura, M., Cascio, D., Desbois, A., Yeates, T.O., Kirilovsky, D., Boussac, A.(2003) Plant Cell Physiol 44: 697-706
- PubMed: 12881497
- DOI: https://doi.org/10.1093/pcp/pcg084
- Primary Citation of Related Structures:
1MZ4 - PubMed Abstract:
First, the crystal structure of cytochrome c-550 (the psbV1 gene product) from the thermophilic cyanobacterium Thermosynechococcus elongatus has been determined to a resolution of 1.8 A. A comparison of the T. elongatus cytochrome c-550 structure to its counterparts from mesophilic organisms, Synechocystis 6803 and Arthrospira maxima, suggests that increased numbers of hydrogen bonds may play a role in the structural basis of thermostability. The cytochrome c-550 in T. elongatus also differs from that in Synechocystis 6803 and Arthrospira maxima in its lack of dimerization and the presence of a trigonal planar molecule, possibly bicarbonate, tightly bound to the heme propionate oxygen atoms. Cytochromes c-550 from T. elongatus, Synechocystis 6803 and Arthrospira maxima exhibit different EPR spectra. A correlation has been done between the heme-axial ligands geometries and the rhombicity calculated from the EPR spectra. This correlation indicates that binding of cytochrome c-550 to Photosystem II is accompanied by structural changes in the heme vicinity. Second, the psbV2 gene product has been found and purified. The UV-visible, EPR and Raman spectra are reported. From the spectroscopic data and from a theoretical structural model based on the cytochrome c-550 structure it is proposed that the 6th ligand of the heme-iron is the Tyr86.
Organizational Affiliation:
Molecular Biology Institute, UCLA, Box 951570, Los Angeles, CA 90095-1570, USA. kerfeld@mbi.ucla.edu