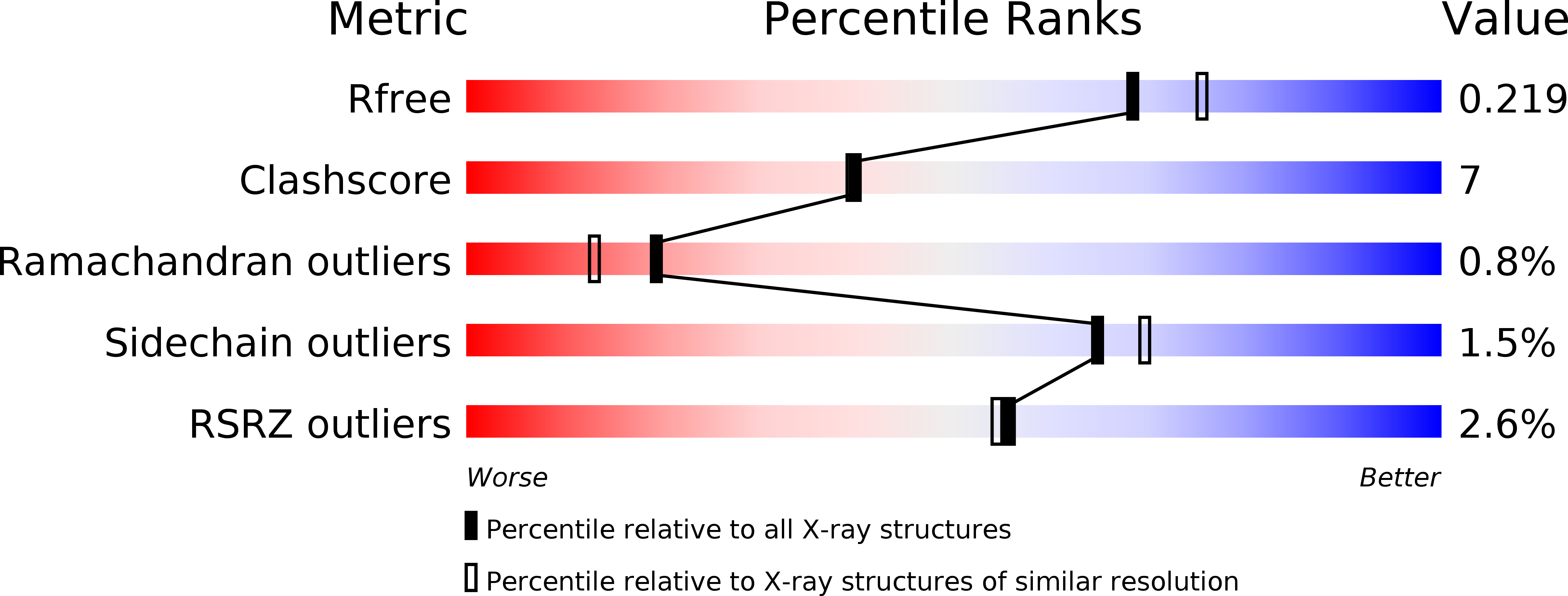
Origins of Protein Stability Revealed by Comparing Crystal Structures of TATA Binding Proteins.
Koike, H., Kawashima-Ohya, Y., Yamasaki, T., Clowney, L., Katsuya, Y., Suzuki, M.(2004) Structure 12: 157-168
- PubMed: 14725775
- DOI: https://doi.org/10.1016/j.str.2003.12.003
- Primary Citation of Related Structures:
1MP9 - PubMed Abstract:
The crystal structure of TATA binding protein (TBP) from a mesothermophilic archaeon, Sulfolobus acidocaldarius, has been determined at a resolution of 2.0 A with an R factor of 20.9%. By comparing this structure with the structures of TBPs from a hyperthermophilic archaeon and mesophilic eukaryotes, as well as by comparing amino acid sequences of TBPs from archaea, covering a wide range of optimum growth temperatures, two significant determinants of the stability of TBP have been identified: increasing the interior hydrophobicity by interaction between three residues, Val, Leu, and Ile, with further differentiation of the surface, and increasing its hydrophilicity and raising the cost of unfolding. These findings suggest directions along which the stability of TBP can be engineered.
Organizational Affiliation:
National Institute of Advanced Industrial Science and Technology, AIST Tsukuba Center 6-10, Higashi 1-1-1, Tsukuba 305-8566, Japan.