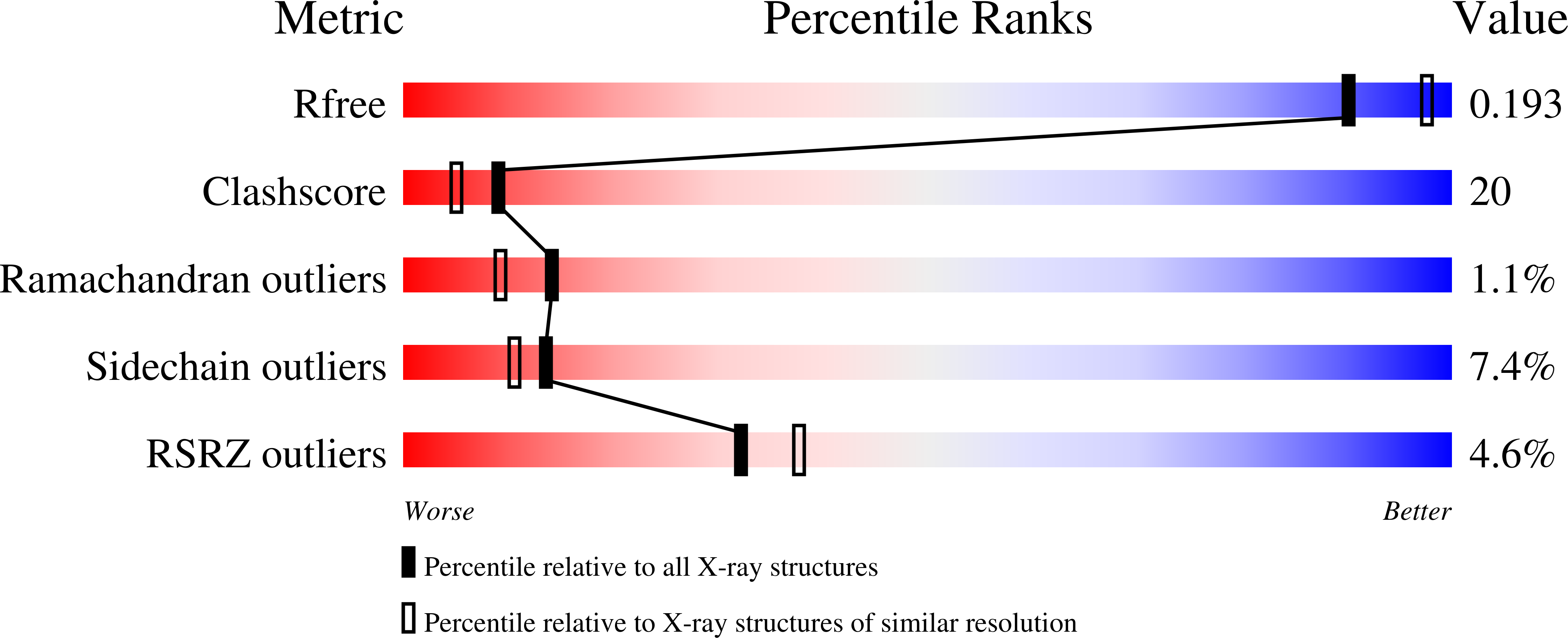
Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Xu, S., Gu, L., Wang, Q., Shu, Y., Song, S., Lin, Z.(2003) Acta Crystallogr D Biol Crystallogr 59: 1574-1581
- PubMed: 12925787
- DOI: https://doi.org/10.1107/s0907444903014598
- Primary Citation of Related Structures:
1M8T - PubMed Abstract:
An acidic PLA(2) (OH APLA(2)-II) from the venom of Ophiophagus hannah (king cobra) shows greater phospholipase A(2) activity and weaker cardiotoxic and myotoxic activity than a homologous acidic PLA(2) from the same venom. The crystal of the enzyme belongs to space group P6(3). The crystals are invariably hemihedrally twinned, exhibiting perfect 622 Laue symmetry. The structure was determined by molecular replacement and refined using a hemihedral twinning program at 2.1 A resolution. The final model has reasonable stereochemistry and a crystallographic R factor of 19.5% (R(free) = 21.5%). The structure reveals the molecular arrangement and the mode of twinning. There are six independent molecules in the asymmetric unit. Owing to the presence of a non-crystallographic twofold parallel to the hemihedral twinning twofold, the molecular packing in the twinned crystal is extremely similar to that in an untwinned crystal for four of the molecules. This unique molecular arrangement may be related to the difficulty in recognizing the twinning. The structure was compared with the previously determined structure of a homologous acidic PLA(2) from the same source. The comparison shows structural changes that might be implicated in the increased catalytic activity and weakened toxicity.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.