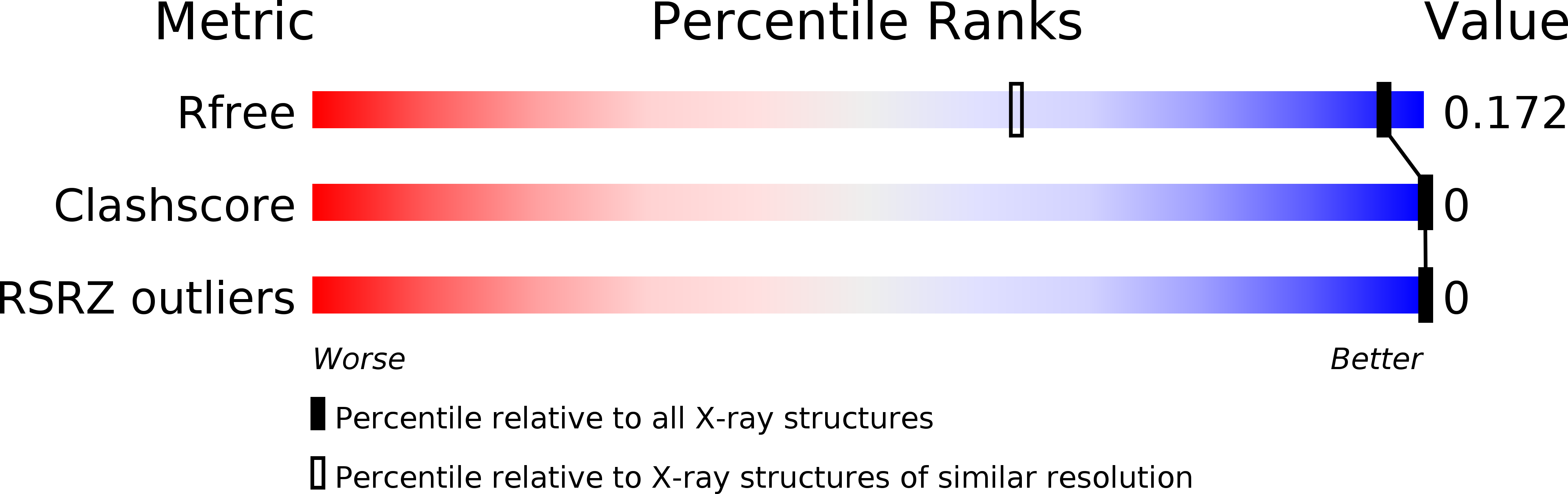Near-atomic resolution crystal structure of an A-DNA decamer d(CCCGATCGGG): cobalt hexammine interaction with A-DNA.
Ramakrishnan, B., Sekharudu, C., Pan, B., Sundaralingam, M.(2003) Acta Crystallogr D Biol Crystallogr 59: 67-72
- PubMed: 12499541
- DOI: https://doi.org/10.1107/s0907444902018917
- Primary Citation of Related Structures:
1M77 - PubMed Abstract:
The structure of the DNA decamer d(CCCGATCGGG) has been determined at 1.25 A resolution. The decamer crystallized in the tetragonal space group P4(3)2(1)2, with unit-cell parameters a = b = 44.3, c = 24.8 A and one strand in the asymmetric unit. The structure was solved by the molecular-replacement method and refined to R(work) and R(free) values of 16.3 and 18.5%, respectively, for 5969 reflections. The decamer forms the A-form DNA duplex, with the abutting crystal packing typical of A-DNA. The crystal packing interactions seem to distort the local conformation: A5 adopts the trans/trans conformation for the torsion angles alpha and gamma instead of the usual gauche(-)/gauche(+) conformations, yielding G*(G.C) base triplets. The highly hydrated [Co(NH(3))(6)](3+) ion adopts a novel binding mode to the DNA duplex, binding directly to phosphate groups and connecting to N7 and O6 atoms of guanines by water bridges. Analysis of thermal parameters (B factors) shows that the nucleotides involved in abutting crystal packing are thermally more stable than other nucleotides in the duplex.
Organizational Affiliation:
Department of Chemistry, The Ohio State University 200 Johnston Laboratory, 176 West 19th Avenue, Columbus, Ohio 43210-1002, USA.