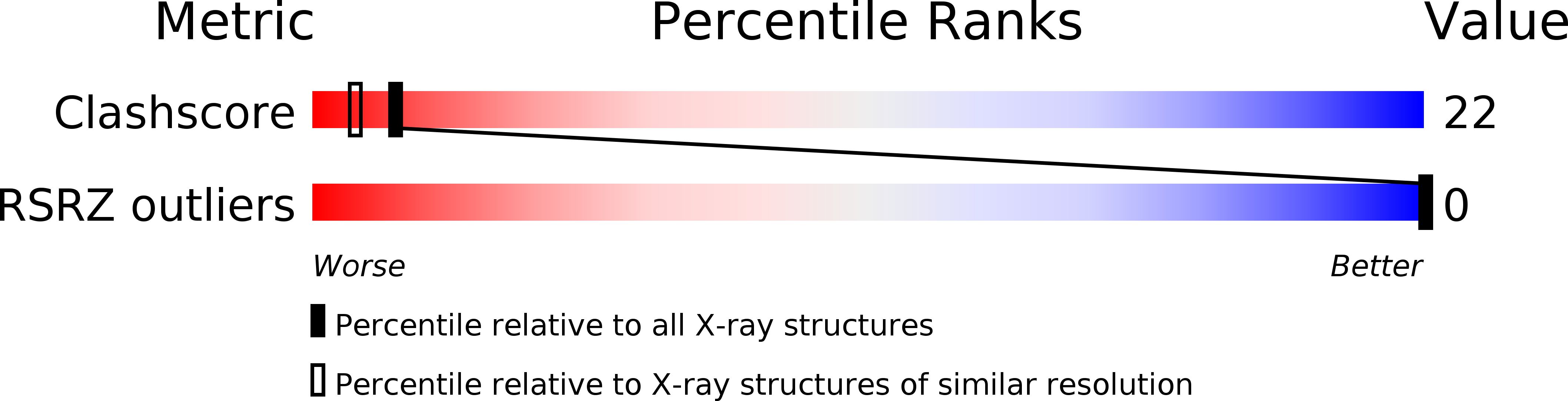Structure of a dicationic monoimidazole lexitropsin bound to DNA.
Goodsell, D.S., Ng, H.L., Kopka, M.L., Lown, J.W., Dickerson, R.E.(1995) Biochemistry 34: 16654-16661
- PubMed: 8527438
- DOI: https://doi.org/10.1021/bi00051a013
- Primary Citation of Related Structures:
1LEX, 1LEY - PubMed Abstract:
An X-ray crystal structure has been solved of the complex of a dicationic lexitropsin with a B-DNA duplex of sequence CGCGAATTCGCG. The lexitropsin is identical to netropsin except for replacement of the first methylpyrrole ring by methylimidazole, converting a =CH- to =N-. Crystals are isomorphous with those of the DNA dodecamer in the absence of drug. Although the =N- for =CH- substitution was intended to make that locus on the drug molecule compatible with a G.C base pair, electrostatic attraction for the two cationic ends of the drug predominates, and this lexitropsin binds to the same central AATT site as does the parent netropsin. But unlike netropsin, this lexitropsin exhibits end-for-end disorder in the crystal. Both orientations were refined separately to completion. Final residual errors at 2.25 A resolution for the 2358 reflections above 2 sigma in F are R = 0.165 for one orientation (LexA) with 37 water molecules and 0.164 for the inverted drug orientation (LexB) with 40 water molecules. This molecular disorder is probably attributable to a weakening of binding to the AATT site occasioned by the imidazole-for-pyrrole substitution.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California at Los Angeles 90095, USA.