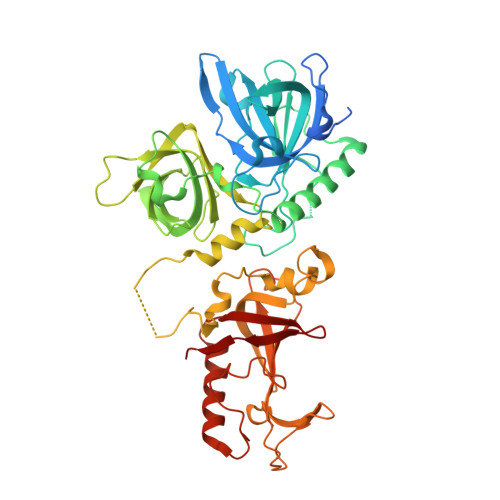
Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA.
Peersen, O.B., Ruggles, J.A., Schultz, S.C.(2002) Nat Struct Biol 9: 182-187
- PubMed: 11836536
- DOI: https://doi.org/10.1038/nsb761
- Primary Citation of Related Structures:
1KIX - PubMed Abstract:
Telomeres are the specialized protein--DNA complexes that cap and protect the ends of linear eukaryotic chromosomes. The extreme 3' end of the telomeric DNA in Oxytricha nova is bound by a two-subunit sequence-specific and 3' end-specific protein called the telomere end-binding protein (OnTEBP). Here we describe the crystal structure of the alpha-subunit of OnTEBP in complex with T4G4 single-stranded telomeric DNA. This structure shows an (alpha--ssDNA)2 homodimer with a large approximately 7,000 A2 protein--protein interface in which the domains of alpha are rearranged extensively from their positions in the structure of an alpha--beta--ssDNA ternary complex. The (alpha--ssDNA)2 complex can bind two telomeres on opposite sides of the dimer and, thus, acts as a protein mediator of telomere--telomere associations. The structures of the (alpha--ssDNA)2 dimer presented here and the previously described alpha--beta--ssDNA complex demonstrate that OnTEBP forms multiple telomeric complexes that potentially mediate the assembly and disassembly of higher order telomeric structures.
Organizational Affiliation:
University of Colorado, Department of Chemistry and Biochemistry, Boulder, Colorado 80309-0215 USA. Olve.Peersen@ColoState.edu