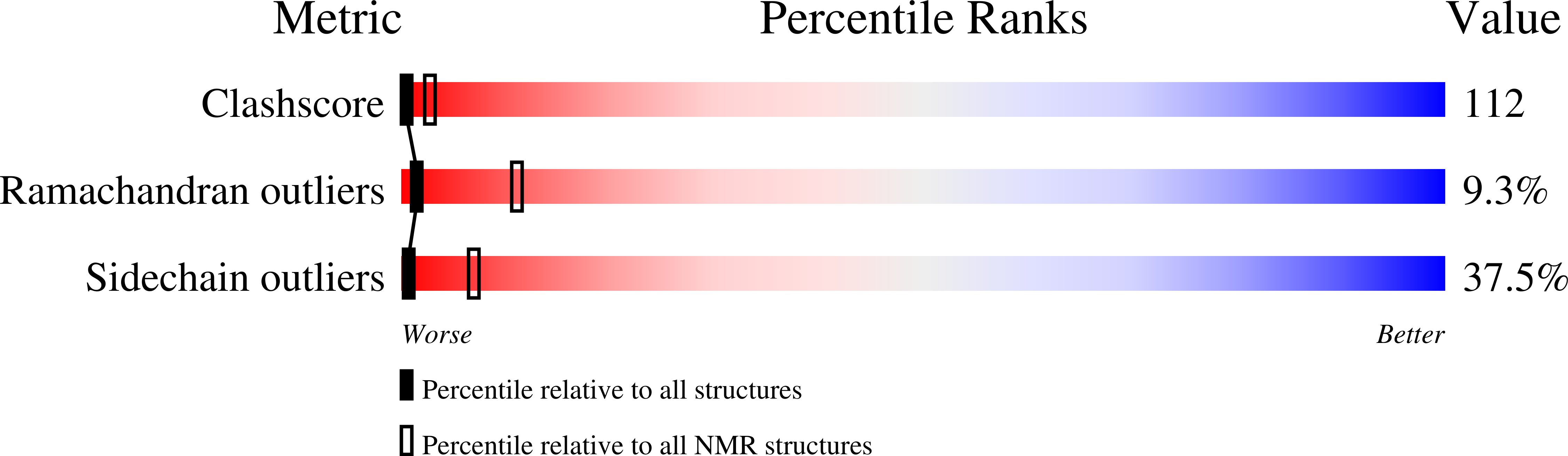Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
Liu, P.P., Chen, Y.C., Li, C., Hsieh, Y.H., Chen, S.W., Chen, S.H., Jeng, W.Y., Chuang, W.J.(2002) Proteins 49: 543-553
- PubMed: 12402362
- DOI: https://doi.org/10.1002/prot.10227
- Primary Citation of Related Structures:
1JXS - PubMed Abstract:
Interleukin enhancer binding factor (ILF) binds to the interleukin-2 (IL-2) promoter and regulates IL-2 gene expression. In this study, the 3D structure of the DNA-binding domain of ILF was determined by multidimensional NMR spectroscopy. NMR structure analysis revealed that the DNA-binding domain of ILF is a new member of the winged helix/forkhead family, and that its wing 2 contains an extra alpha-helix. This is the first study to report the presence of a C-terminal alpha-helix in place of a typical wing 2 in a member of this family. This structural difference may be responsible for the different DNA-binding specificity of ILF compared to other winged helix/forkhead proteins. Our deletion studies of the fragments of ILF also suggest that the C-terminal region plays a regulatory role in DNA binding.
Organizational Affiliation:
Department of Biochemistry, National Cheng Kung University College of Medicine, Tainan 701, Taiwan.