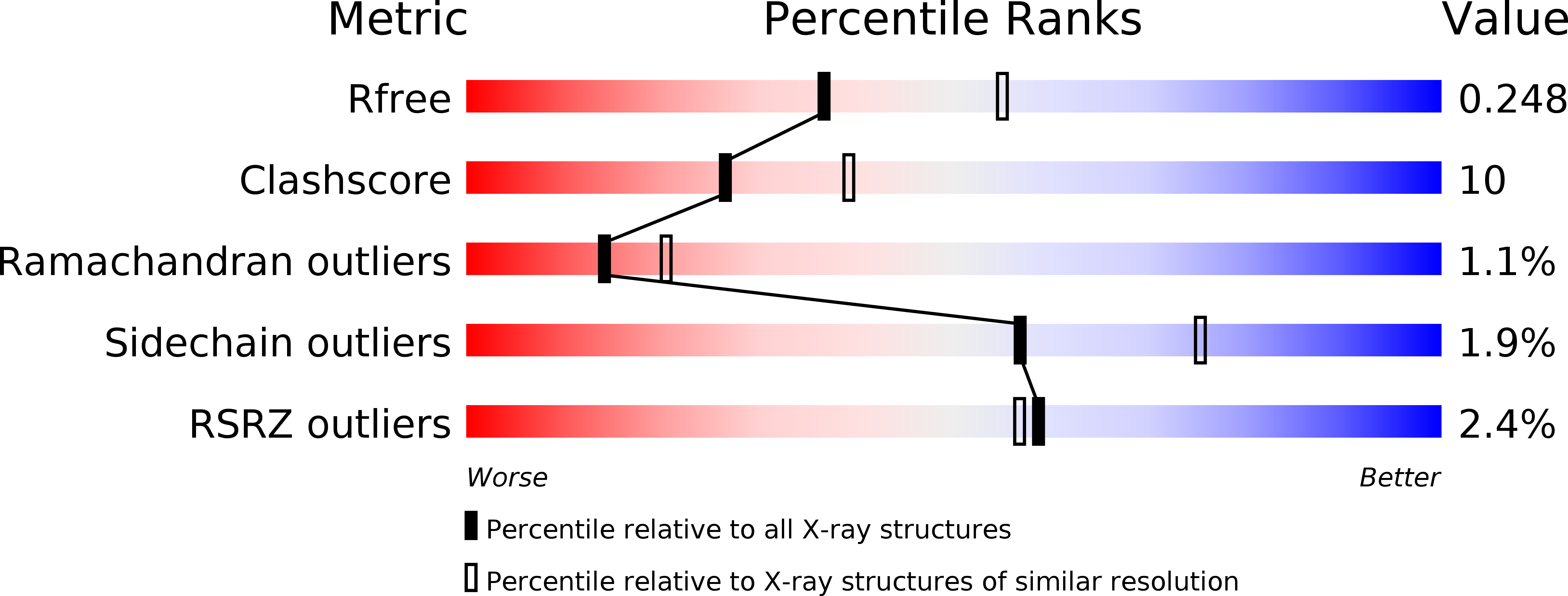
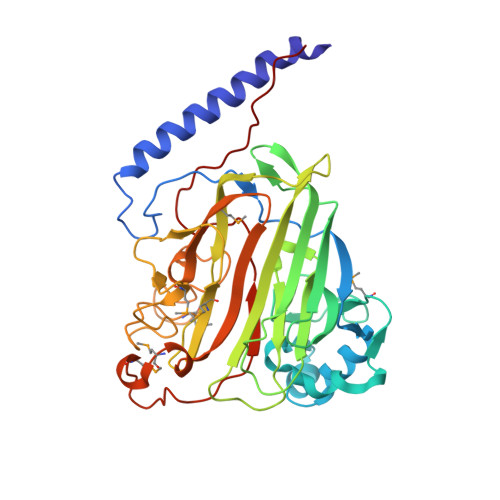
Crystal Structure of the V-region of Streptococcus mutans Antigen I/II at 2.4 a Resolution Suggests a Sugar Preformed Binding Site
Troffer-Charlier, N., Ogier, J., Moras, D., Cavarelli, J.(2002) J Mol Biol 318: 179-188
- PubMed: 12054777
- DOI: https://doi.org/10.1016/S0022-2836(02)00025-6
- Primary Citation of Related Structures:
1JMM - PubMed Abstract:
Antigens I/II are large multifunctional adhesins from oral viridans streptococci that exert immunomodulatory effects on human cells and play important roles in inflammatory disorders. Among them, Streptococcus mutans plays a major role in the initiation of dental caries. The structure of the V-region (SrV+, residues 464-840) of the antigen I/II of S. mutans has been determined using the multiwavelength anomalous diffraction phasing technique with seleno-methionine-substituted recombinant protein and subsequently refined at 2.4 A resolution. The crystal structure of SrV+ revealed a lectin-like fold that displays a putative preformed carbohydrate-binding site stabilized by a metal ion. Inhibition of this binding site may confer to humans a protection against dental caries and dissemination of the bacteria to extra-oral sites involved in life-threatening inflammatory diseases. This crystal structure constitutes a first step in understanding the structure-function relationship of antigens I/II and may help in delineating new preventive or therapeutic strategies against colonization of the host by oral streptococci.
Organizational Affiliation:
Institut National de la Santé et de la Recherche Médicale U424, Faculté de Chirurgie Dentaire, Université Louis Pasteur, 11 rue Humann, 67085 Strasbourg, France.