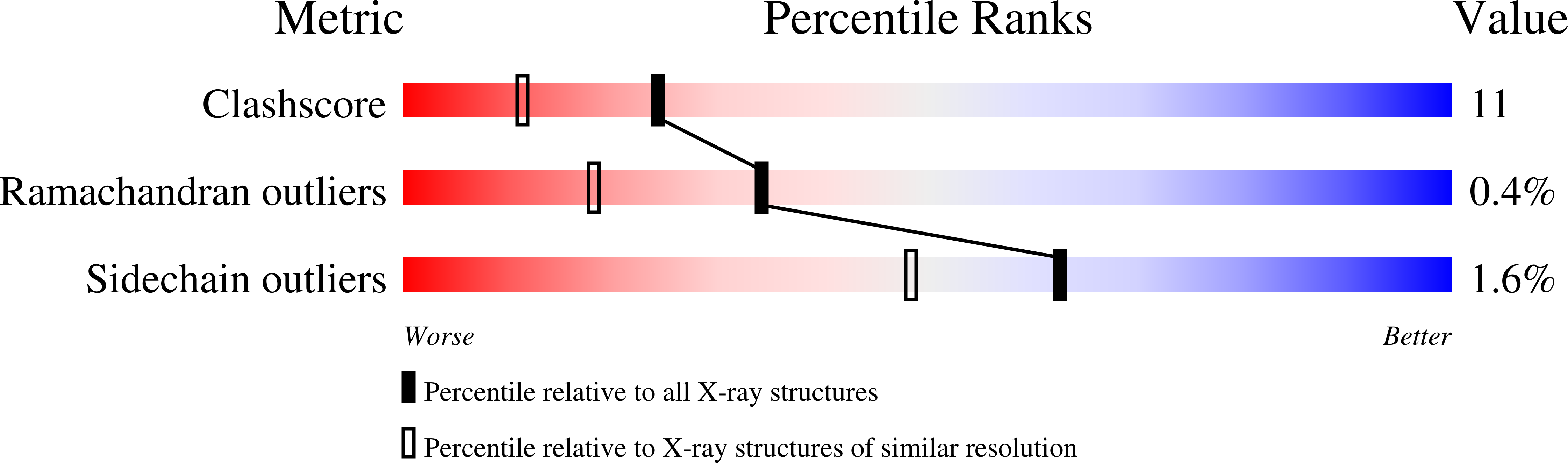X-ray structure of Saccharomyces cerevisiae homologous mitochondrial matrix factor 1 (Hmf1).
Deaconescu, A.M., Roll-Mecak, A., Bonanno, J.B., Gerchman, S.E., Kycia, H., Studier, F.W., Burley, S.K.(2002) Proteins 48: 431-436
- PubMed: 12112709
- DOI: https://doi.org/10.1002/prot.10151
- Primary Citation of Related Structures:
1JD1
Organizational Affiliation:
Laboratories of Molecular Biophysics, The Rockefeller University, New York, New York, USA.