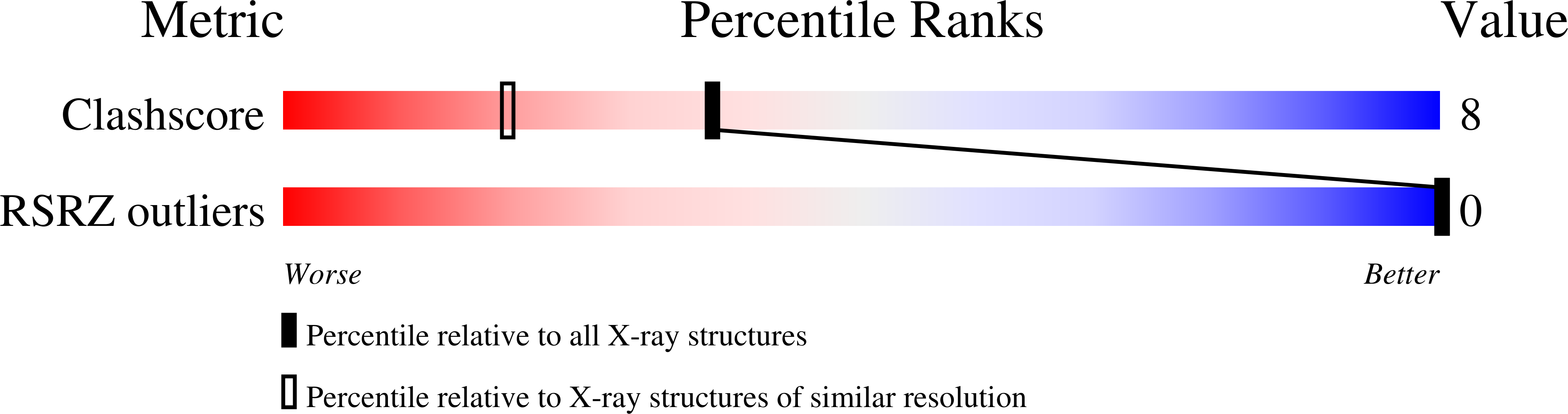Crystallographic studies on damaged DNAs IV. N(4)-methoxycytosine shows a second face for Watson-Crick base-pairing, leading to purine transition mutagenesis.
Hossain, M.T., Sunami, T., Tsunoda, M., Hikima, T., Chatake, T., Ueno, Y., Matsuda, A., Takenaka, A.(2001) Nucleic Acids Res 29: 3949-3954
- PubMed: 11574676
- DOI: https://doi.org/10.1093/nar/29.19.3949
- Primary Citation of Related Structures:
1J8L - PubMed Abstract:
To investigate the mutation mechanism of purine transitions in DNA damaged with methoxyamine, a DNA dodecamer with the sequence d(CGCAAATTmo(4)CGCG), where mo(4)C is 2'-deoxy-N(4)-methoxycytidine, has been synthesized and the crystal structure determined by X-ray analysis. The duplex structure is similar to that of the original undamaged B-form dodecamer, indicating that the methoxylation does not affect the overall DNA conformation. Electron density maps clearly show that the two mo(4)C residues form Watson-Crick-type base pairs with the adenine residues of the opposite strand and that the methoxy groups of mo(4)C adopt the anti conformation to N(3) around the C(4)-N(4) bond. For the pair formation through hydrogen bonds the mo(4)C residues are in the imino tautomeric state. Together with previous work, the present work establishes that the methoxylated cytosine residue can present two alternate faces for Watson-Crick base-pairing, thanks to the amino<-->imino tautomerism allowed by methoxylation. Based on this property, two gene transition routes are proposed.
Organizational Affiliation:
Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Nagatsuta, Midori-ku, Yokohama 226-8501, Japan.