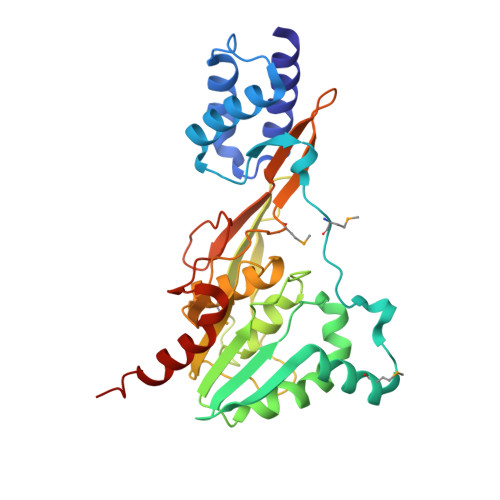
Crystal structure of the functional unit of interphotoreceptor retinoid binding protein.
Loew, A., Gonzalez-Fernandez, F.(2002) Structure 10: 43-49
- PubMed: 11796109
- DOI: https://doi.org/10.1016/s0969-2126(01)00698-0
- Primary Citation of Related Structures:
1J7X - PubMed Abstract:
Interphotoreceptor retinoid binding protein (IRBP), the major soluble component of the interphotoreceptor matrix, is critical to the function, integrity, and development of the vertebrate retina. Although its role is poorly understood, IRBP has been thought to protect 11-cis retinal and all-trans retinol while facilitating their exchange between the photoreceptors and retinal-pigmented epithelium. We determined the X-ray structure of one of the functional units, or modules, of Xenopus laevis IRBP to 1.8 A resolution by multiwavelength anomalous dispersion. The monomeric protein consists of two domains separated by a hydrophobic ligand binding site. A structural homology to the recently solved photosystem II D1 C-terminal-processing protease and the enoyl-CoA isomerase/hydratase family suggests the utility of a common fold used in diverse settings, ranging from proteolysis to fatty acid isomerization to retinoid transport.
Organizational Affiliation:
Department of Biochemistry, University of Texas, Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, TX 75390, USA. andreas.loew@abbott.com