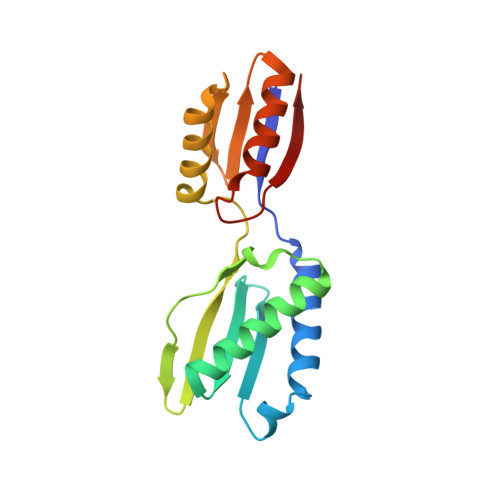
Crystal structure of the YajQ protein from Haemophilus influenzae reveals a tandem of RNP-like domains
Teplyakov, A., Obmolova, G., Bir, N., Reddy, P., Howard, A.J., Gilliland, G.L.(2003) J Struct Funct Genomics 4: 1-9
- PubMed: 12943362
- DOI: https://doi.org/10.1023/a:1024620416876
- Primary Citation of Related Structures:
1IN0 - PubMed Abstract:
A hypothetical protein encoded by the gene YajQ of Haemophilus influenzae was selected, as part of a structural genomics project, for X-ray crystallographic structure determination and analysis to assist with the functional assignment. The protein is present in most bacteria, but not in archaea or eukaryotes. The amino acid sequence has no homology to that of other proteins. The YajQ protein was cloned, expressed, and the crystal structure determined at 2.1-A resolution by applying the multiwavelength anomalous dispersion method to a mercury derivative. The polypeptide chain is folded into two domains with identical folding topology. Each domain has a four-stranded antiparallel beta-sheet flanked on one side by two alpha-helices. This structural motif is a characteristic feature of many RNA-binding proteins. The tetrameric structure observed in the crystal suggests a possibility of binding two stretches of double-stranded nucleic acid.
Organizational Affiliation:
Center for Advanced Research in Biotechnology of the University of Maryland Biotechnology Institute and the National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA. alexey@carb.nist.gov