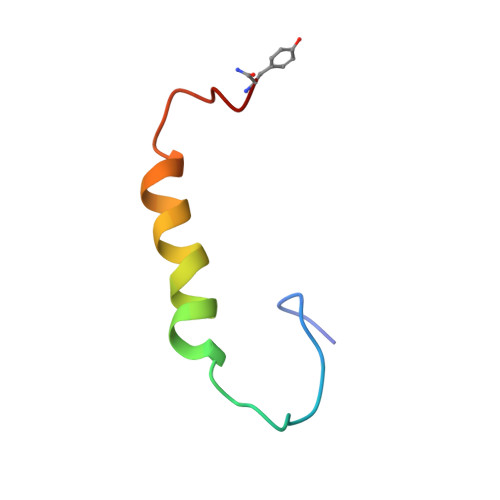
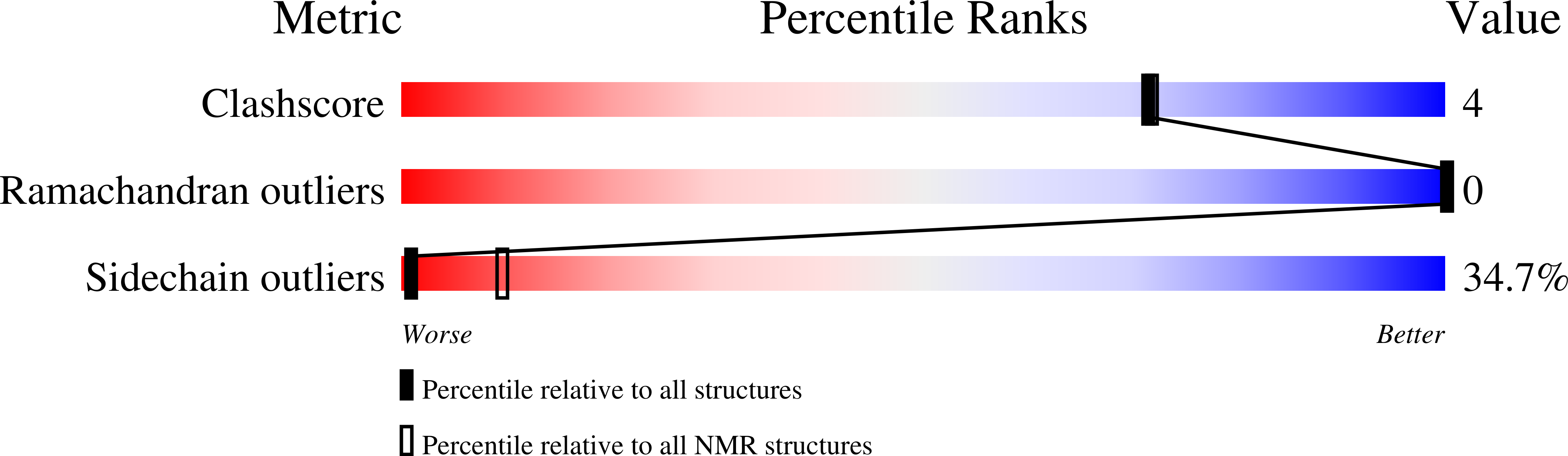Key motif to gain selectivity at the neuropeptide Y5-receptor: structure and dynamics of micelle-bound [Ala31, Pro32]-NPY.
Bader, R., Rytz, G., Lerch, M., Beck-Sickinger, A.G., Zerbe, O.(2002) Biochemistry 41: 8031-8042
- PubMed: 12069594
- DOI: https://doi.org/10.1021/bi0201419
- Primary Citation of Related Structures:
1ICY - PubMed Abstract:
The structure of [Ala(31), Pro(32)]-NPY, a neuropeptide Y mutant with selectivity for the NPY Y(5)-receptor (Cabrele, C., Wieland, H. A., Stidsen, C., Beck-Sickinger, A. G., (2002) Biochemistry XX, XXXX-XXXX (companion paper)), has been characterized in the presence of the membrane mimetic dodecylphosphocholine (DPC) micelles using high-resolution NMR techniques. The overall topology closely resembles the fold of the previously described Y(5)-receptor-selective agonist [Ala(31), Aib(32)]-NPY (Cabrele, C., Langer, M., Bader, R., Wieland, H. A., Doods, H. N., Zerbe, O., and Beck-Sickinger, A. G. (2000) J. Biol. Chem 275, 36043-36048). Similar to wild-type neuropeptide Y (NPY) and [Ala(31), Aib(32)]-NPY, the N-terminal residues Tyr(1)-Asp(16) are disordered in solution. Starting from residue Leu(17), an alpha helix extends toward the C-terminus. The decreased density of medium-range NOEs for the C-terminal residues resulting in larger RMSD values for the backbone atoms of Ala(31)-Tyr(36) indicates that the alpha helix has become interrupted through the [Ala(31), Pro(32)] mutation. This finding is further supported by (15)N-relaxation data through which we can demonstrate that the well-defined alpha helix is restricted to residues 17-31, with the C-terminal tetrapeptide displaying increased flexibility as compared to NPY. Surprisingly, increased generalized order parameter as well as decreased (3)J(HN)(alpha) scalar coupling constants reveal that the central helix is stabilized in comparison to wild-type NPY. Micelle-integrating spin labels were used to probe the mode of association of the helix with the membrane mimetic. The Y(5)-receptor-selective mutant and NPY share a similar orientation, which is parallel to the lipid surface. However, signal reductions due to efficient electron, nuclear spin relaxation were much less pronounced for the surface-averted residues in [Ala(31), Pro(32)]-NPY when compared to wild-type DPC-bound NPY. Only the signals of residues Asn(29) and Leu(30) were significantly more reduced in the mutant. The postulation of a different membrane binding mode of [Ala(31), Pro(32)]-NPY is further supported by the faster H/D exchange at the C-terminal amide protons. We conclude that arginine residues 33 and 35, which are believed to be directly involved in forming contacts to acidic receptor residues at the membrane-water interface, are no longer fixed in a well-defined conformation close to the membrane surface in [Ala(31), Pro(32)]-NPY.
Organizational Affiliation:
Institute of Pharmaceutical Sciences, ETH Zürich, Winterthurerstrasse 190, 8057 Zürich, Switzerland.