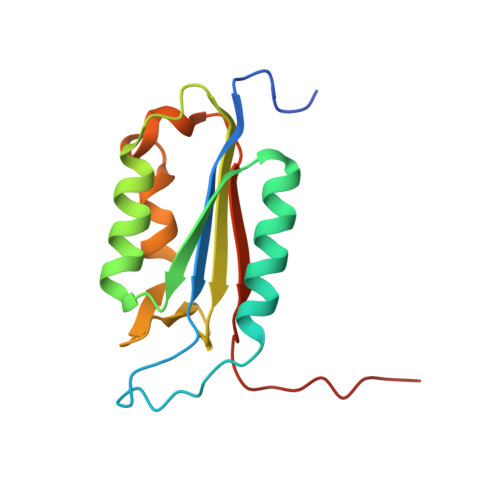
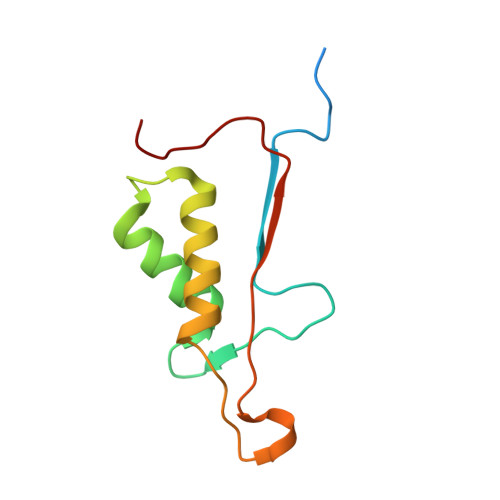
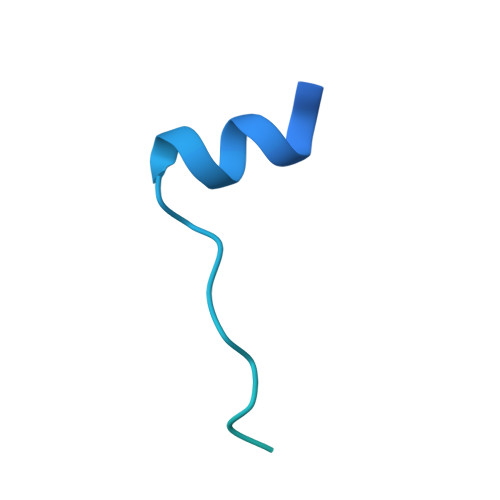
Structural basis of caspase-7 inhibition by XIAP.
Chai, J., Shiozaki, E., Srinivasula, S.M., Wu, Q., Datta, P., Alnemri, E.S., Shi, Y., Dataa, P.(2001) Cell 104: 769-780
- PubMed: 11257230
- DOI: https://doi.org/10.1016/s0092-8674(01)00272-0
- Primary Citation of Related Structures:
1I51 - PubMed Abstract:
The inhibitor of apoptosis (IAP) proteins suppress cell death by inhibiting the catalytic activity of caspases. Here we present the crystal structure of caspase-7 in complex with a potent inhibitory fragment from XIAP at 2.45 A resolution. An 18-residue XIAP peptide binds the catalytic groove of caspase-7, making extensive contacts to the residues that are essential for its catalytic activity. Strikingly, despite a reversal of relative orientation, a subset of interactions between caspase-7 and XIAP closely resemble those between caspase-7 and its tetrapeptide inhibitor DEVD-CHO. Our biochemical and structural analyses reveal that the BIR domains are dispensable for the inhibition of caspase-3 and -7. This study provides a structural basis for the design of the next-generation caspase inhibitors.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.