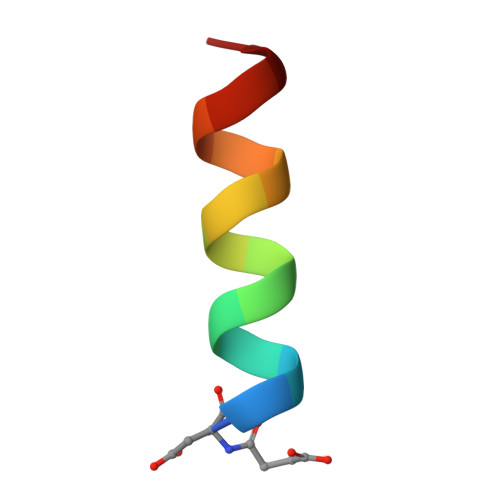
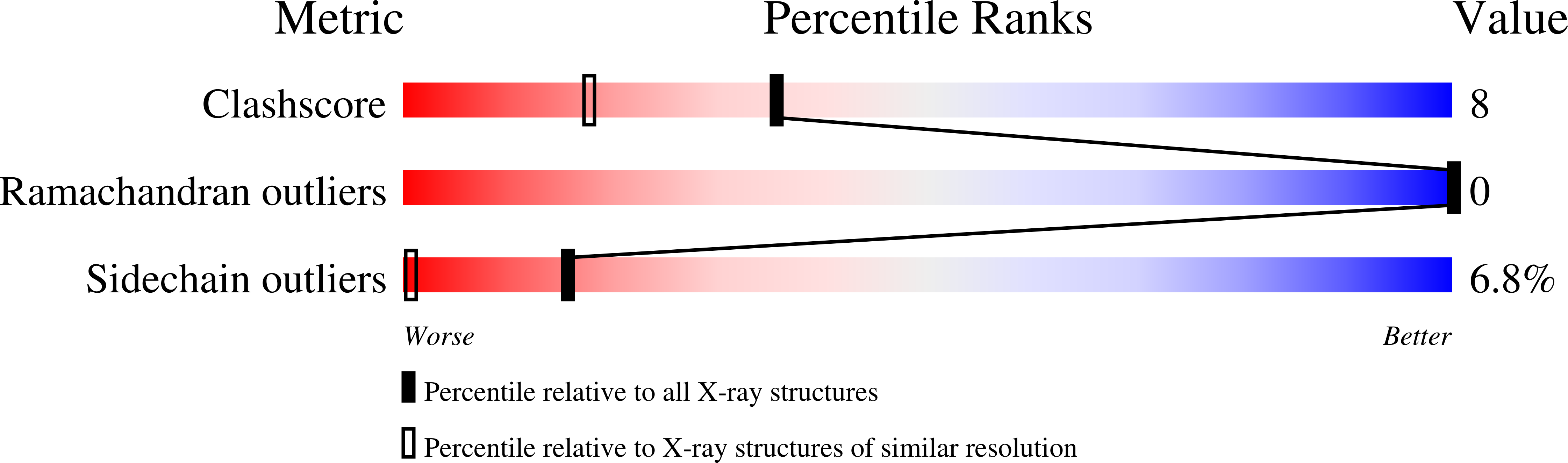Design of a minimal protein oligomerization domain by a structural approach.
Burkhard, P., Meier, M., Lustig, A.(2000) Protein Sci 9: 2294-2301
- PubMed: 11206050
- DOI: https://doi.org/10.1110/ps.9.12.2294
- Primary Citation of Related Structures:
1HQJ - PubMed Abstract:
Because of the simplicity and regularity of the alpha-helical coiled coil relative to other structural motifs, it can be conveniently used to clarify the molecular interactions responsible for protein folding and stability. Here we describe the de novo design and characterization of a two heptad-repeat peptide stabilized by a complex network of inter- and intrahelical salt bridges. Circular dichroism spectroscopy and analytical ultracentrifugation show that this peptide is highly alpha-helical and 100% dimeric tinder physiological buffer conditions. Interestingly, the peptide was shown to switch its oligomerization state from a dimer to a trimer upon increasing ionic strength. The correctness of the rational design principles used here is supported by details of the atomic structure of the peptide deduced from X-ray crystallography. The structure of the peptide shows that it is not a molten globule but assumes a unique, native-like conformation. This de novo peptide thus represents an attractive model system for the design of a molecular recognition system.
Organizational Affiliation:
M.E. Muller Institute for Structural Biology, Biozentrum, University of Basel, Switzerland.