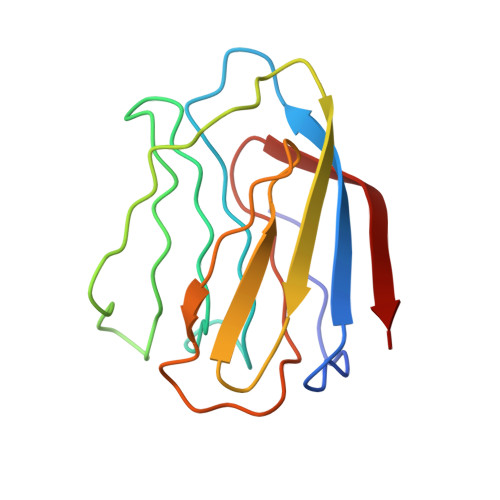
X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution.
Lobsanov, Y.D., Gitt, M.A., Leffler, H., Barondes, S.H., Rini, J.M.(1993) J Biol Chem 268: 27034-27038
- PubMed: 8262940
- DOI: https://doi.org/10.2210/pdb1hlc/pdb
- Primary Citation of Related Structures:
1HLC - PubMed Abstract:
S-Lac lectins are a family of soluble lactose-binding animal lectins, some of which have been implicated in modulating cell-cell and cell-matrix interactions through specific carbohydrate-mediated recognition. We report here the x-ray crystal structure of a representative member of this family, the human dimeric S-Lac lectin, L-14-II, in complex with lactose, at 2.9-A resolution. The two-fold symmetric dimer is made up of two extended anti-parallel beta-sheets, which associate in a beta-sandwich motif. Remarkably, the L-14-II monomer shares not only the same topology, but a very similar beta-sheet structure with that of the leguminous plant lectins, suggesting a conserved structure-function relationship. Carbohydrate binding by L-14-II was found to involve protein residues that are very highly conserved among all S-Lac lectins. These residues map to a single DNA exon, suggesting a carbohydrate binding cassette common to all S-Lac lectins.
Organizational Affiliation:
Department of Molecular and Medical Genetics, University of Toronto, Ontario, Canada.