A new structural class of serine protease inhibitors revealed by the structure of the hirustasin-kallikrein complex.
Mittl, P.R., Di Marco, S., Fendrich, G., Pohlig, G., Heim, J., Sommerhoff, C., Fritz, H., Priestle, J.P., Grutter, M.G.(1997) Structure 5: 253-264
- PubMed: 9032072
- DOI: https://doi.org/10.1016/s0969-2126(97)00183-4
- Primary Citation of Related Structures:
1HIA - PubMed Abstract:
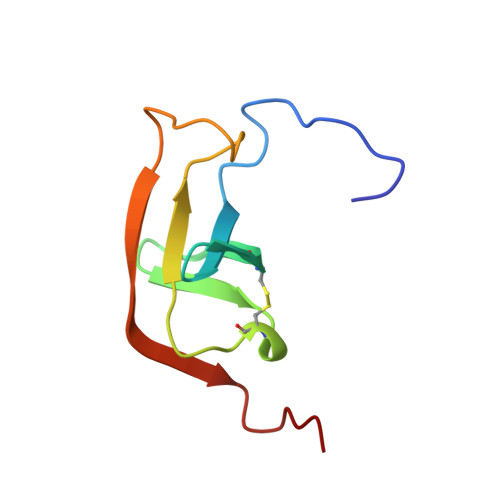
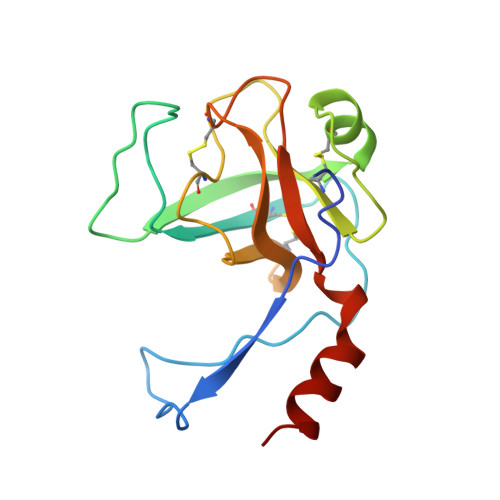
Hirustasin belongs to a class of serine protease inhibitors characterized by a well conserved pattern of cysteine residues. Unlike the closely related inhibitors, antistasin/ghilanten and guamerin, which are selective for coagulation factor Xa or neutrophil elastase, hirustasin binds specifically to tissue kallikrein. The conservation of the pattern of cysteine residues and the significant sequence homology suggest that these related inhibitors possess a similar three-dimensional structure to hirustasin. The crystal structure of the complex between tissue kallikrein and hirustasin was analyzed at 2.4 resolution. Hirustasin folds into a brick-like structure that is dominated by five disulfide bridges and is sparse in secondary structural elements. The cysteine residues are connected in an abab cdecde pattern that causes the polypeptide chain to fold into two similar motifs. As a hydrophobic core is absent from hirustasin the disulfide bridges maintain the tertiary structure and present the primary binding loop to the active site of the protease. The general structural topography and disulfide connectivity of hirustasin has not previously been described. The crystal structure of the kallikrein-hirustasin complex reveals that hirustasin differs from other serine protease inhibitors in its conformation and its disulfide bond connectivity, making it the prototype for a new class of inhibitor. The disulfide pattern shows that the structure consists of two domains, but only the C-terminal domain interacts with the protease. The disulfide pattern of the N-terminal domain is related to the pattern found in other proteins. Kallikrein recognizes hirustasin by the formation of an antiparallel beta sheet between the protease and the inhibitor. The P1 arginine binds in a deep negatively charged pocket of the enzyme. An additional pocket at the periphery of the active site accommodates the sidechain of the P4 valine.
Organizational Affiliation:
CDDT Pharmaceutical Research, Ciba-Geigy Ltd., 4002 Basel, Switzerland.