Structure of Cytochrome C7 from Desulfuromonas Acetoxidans at 1.9A Resolutio N
Czjzek, M., Arnoux, P., Haser, R., Shepard, W.(2001) Acta Crystallogr D Biol Crystallogr 57: 670
- PubMed: 11320307
- DOI: https://doi.org/10.1107/s0907444901003481
- Primary Citation of Related Structures:
1HH5 - PubMed Abstract:
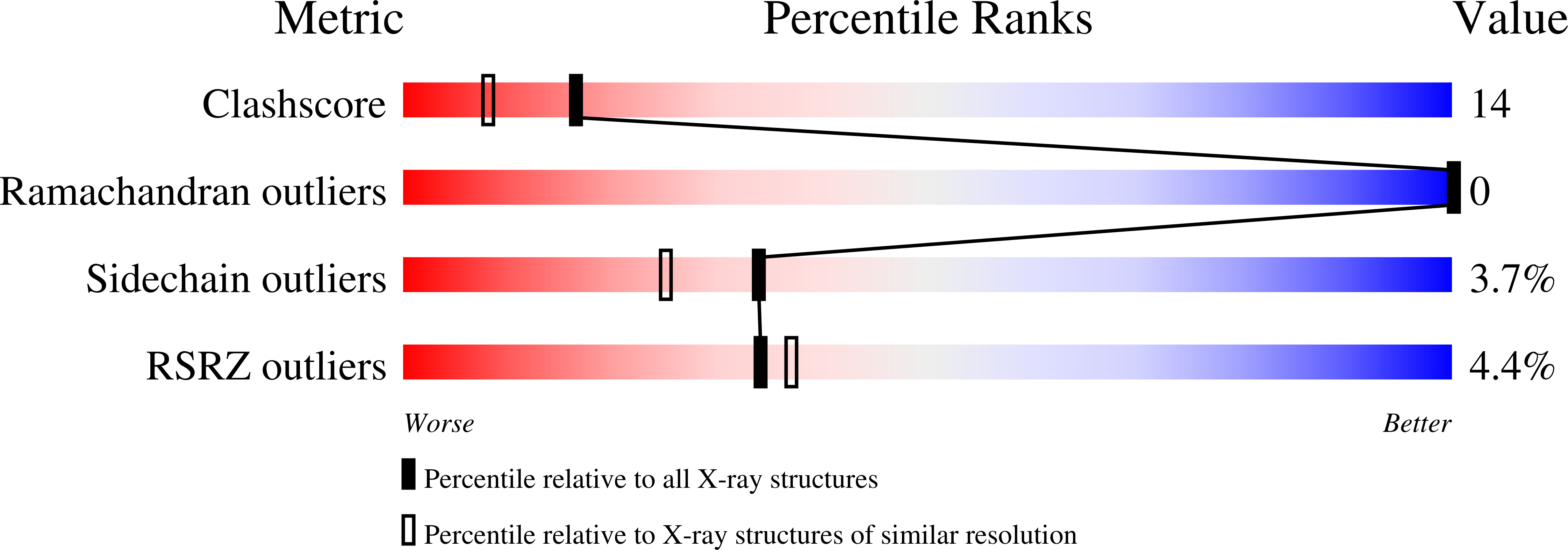
Multihaem cytochromes play a key role in electron-transport reactions in the periplasm of sulfate- and sulfur-reducing bacteria. The redox proteins grouped in the c3 superfamily also display metal-reducing activities, which make them interesting biotechnological tools. The crystal structure of the fully oxidized cytochrome c7 from Desulfuromonas acetoxidans has been solved by combined molecular-replacement and MAD methods. The structure has been refined at 1.9 A resolution to an R value of 19.1% (R(free) = 24.3%) and includes three haems and 116 water molecules. The protein displays the cytochrome c3 fold in a highly minimized form, while haem 2 and the surrounding protein environment are missing. The geometry of haem packing and of the haem axial ligands and propionates are described and compared with that of c3 cytochromes. The crystal structure is compared with the solution structure recently obtained by NMR methods and with its homologue cytochromes of the c3 superfamily. Comparison of the high number of available structures makes it possible to analyze the structural role of the few highly conserved residues, in addition to the cysteines and histidines that link the porphyrin rings and the Fe atoms to the protein chain.
Organizational Affiliation:
Laboratoire d'Architecture et de Fonction des Macromolécules Biologiques, CNRS et Université Aix Marseille I and II, Marseille, 31 Chemin Joseph-Aiguer, 13402 Marseille CEDEX 20, France. czjzek@afmb.cnrs-mrs.fr