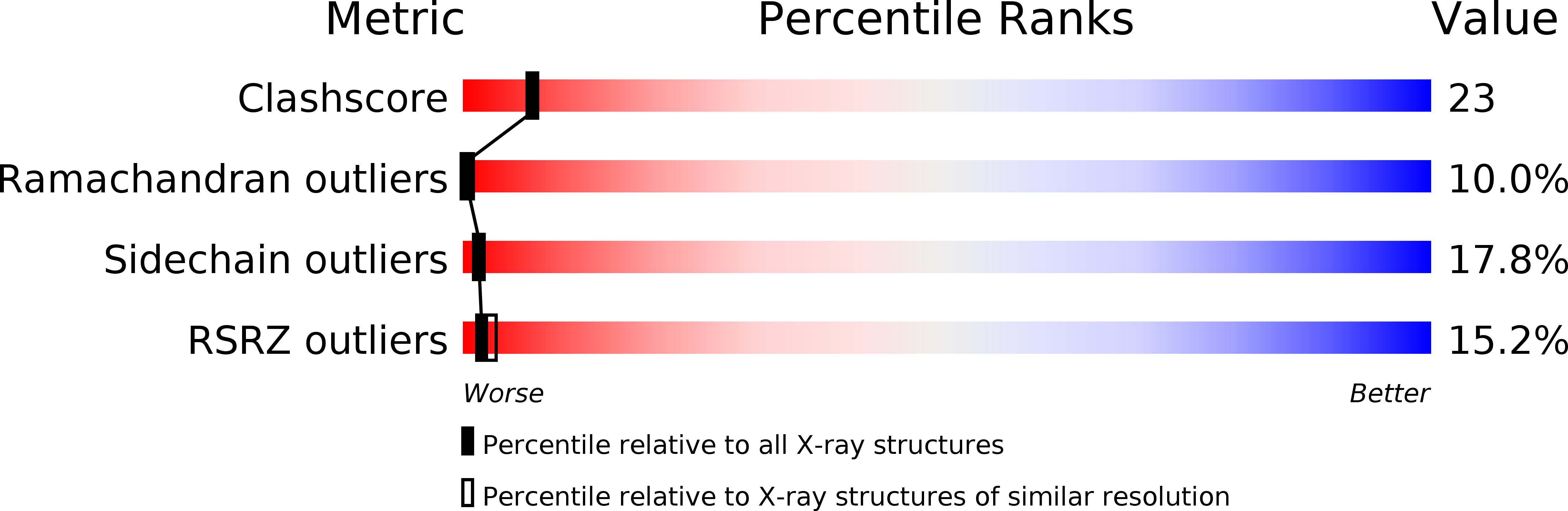Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions.
Feng, J.A., Johnson, R.C., Dickerson, R.E.(1994) Science 263: 348-355
- PubMed: 8278807
- DOI: https://doi.org/10.1126/science.8278807
- Primary Citation of Related Structures:
1HCR - PubMed Abstract:
The structure of the 52-amino acid DNA-binding domain of the prokaryotic Hin recombinase, complexed with a DNA recombination half-site, has been solved by x-ray crystallography at 2.3 angstrom resolution. The Hin domain consists of a three-alpha-helix bundle, with the carboxyl-terminal helix inserted into the major groove of DNA, and two flanking extended polypeptide chains that contact bases in the minor groove. The overall structure displays features resembling both a prototypical bacterial helix-turn-helix and the eukaryotic homeodomain, and in many respects is an intermediate between these two DNA-binding motifs. In addition, a new structural motif is seen: the six-amino acid carboxyl-terminal peptide of the Hin domain runs along the minor groove at the edge of the recombination site, with the peptide backbone facing the floor of the groove and side chains extending away toward the exterior. The x-ray structure provides an almost complete explanation for DNA mutant binding studies in the Hin system and for DNA specificity observed in the Hin-related family of DNA invertases.
Organizational Affiliation:
Molecular Biology Institute, University of California, Los Angeles 90024.