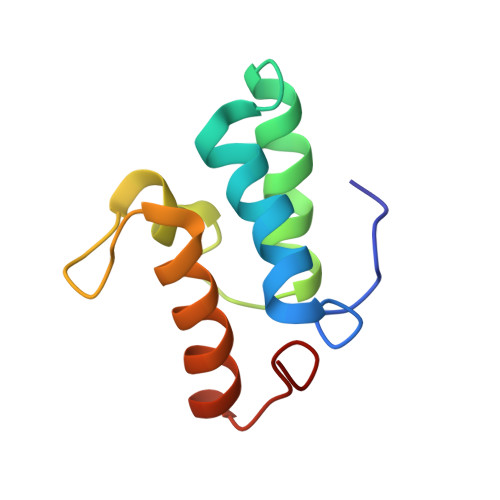
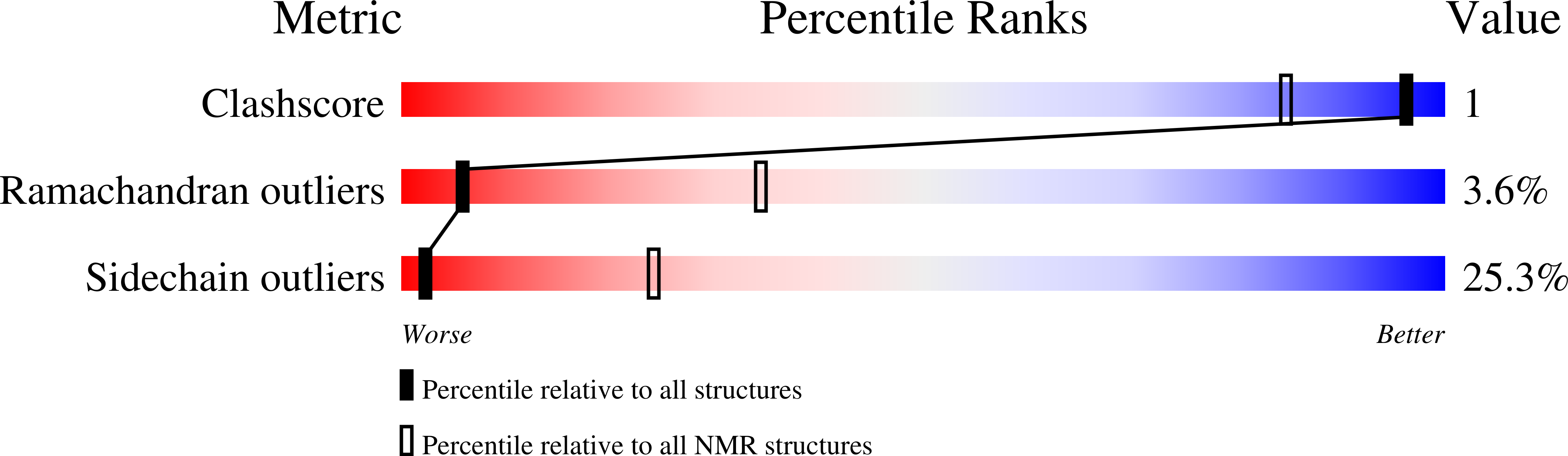Non-Cognate Protein-Protein Interaction: The NMR Structure of the Colicin E8 Inhibitor Protein Im8 and its Interaction with the DNase Domain of Colicin E9
Le Duff, C.S., Videler, H., Boetzel, R., Czisch, M., James, R., Kleanthous, C., Moore, G.R.To be published.