Structure and dynamics of the human granulocyte colony-stimulating factor determined by NMR spectroscopy. Loop mobility in a four-helix-bundle protein.
Zink, T., Ross, A., Luers, K., Cieslar, C., Rudolph, R., Holak, T.A.(1994) Biochemistry 33: 8453-8463
- PubMed: 7518249
- DOI: https://doi.org/10.1021/bi00194a009
- Primary Citation of Related Structures:
1GNC - PubMed Abstract:
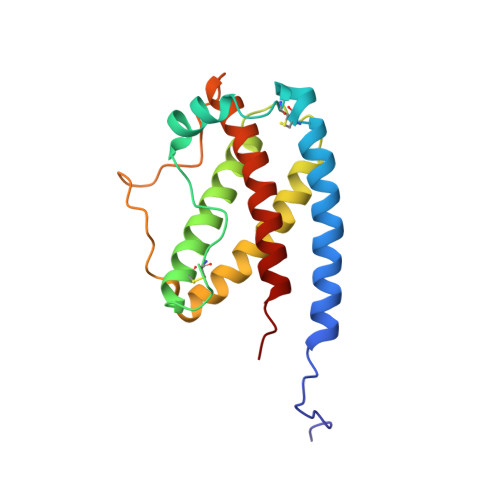
Recombinant 15N- and 13C-labeled human granulocyte colony-stimulating factor (rh-metG-CSF) has been studied by 2D and 3D NMR using uniformly labeled protein, as well as residue-specific 15N-labeled samples. Assignment of 90% of the backbone resonances and 85% of side-chain resonances has enabled the determination of both the secondary and tertiary structures of the protein. The fold is similar to those of the human growth hormone and other growth factors. Four stretches of helices were identified between residues 11 and 41 (helix A), 71 and 95 (helix B), 102 and 125 (helix C), and 145 and 170 (helix D), which form a left-handed four-helix bundle with helices A and B aligned parallel to one another (up-up) and antiparallel to helices C and D (down-down). An additional short fifth helix (E) is part of the AB loop connecting helices A and B. Examination of the protein's relaxation behavior, based on the model-free approach of Lipari and Szabo, shows that the G-CSF backbone has a well-defined structure of limited conformational flexibility in helices. In contrast, the long loop connecting helices C and D exhibits substantial fast internal motion compared to the overall rotational correlation time of the whole molecule, which is on the order of 13 ns.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Martinsried, F.R.G.