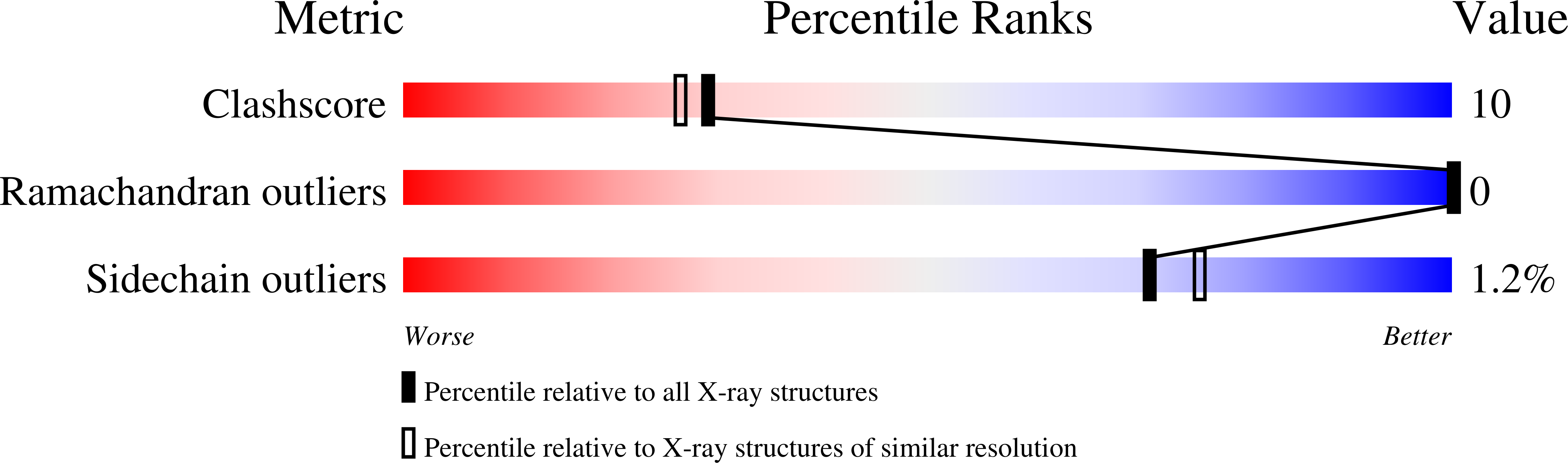A helical lid converts a sulfotransferase to a dehydratase.
Pakhomova, S., Kobayashi, M., Buck, J., Newcomer, M.E.(2001) Nat Struct Biol 8: 447-451
- PubMed: 11323722
- DOI: https://doi.org/10.1038/87617
- Primary Citation of Related Structures:
1FMJ, 1FML - PubMed Abstract:
We report here the crystal structure of retinol dehydratase, an enzyme that catalyzes the synthesis of anhydroretinol. The enzyme is a member of the sulfotransferase superfamily and its crystal structure reveals the insertion of a helical lid into a canonical sulfotransferase fold. Site-directed mutations demonstrate that this inserted lid is necessary for anhydroretinol production but not for sulfonation; thus, insertion of a helical lid can convert a sulfotransferase into a dehydratase.
Organizational Affiliation:
Biochemistry Department, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.