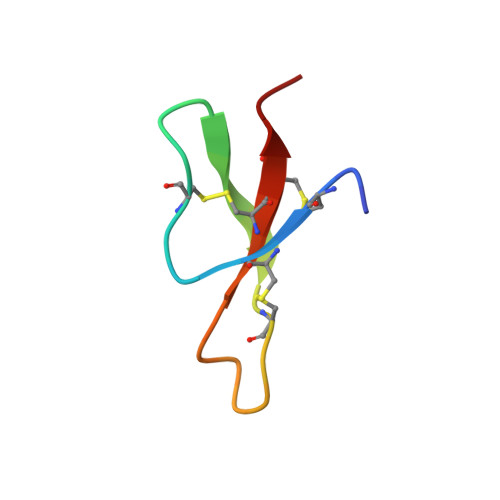
Three-dimensional structure of RK-1: a novel alpha-defensin peptide.
McManus, A.M., Dawson, N.F., Wade, J.D., Carrington, L.E., Winzor, D.J., Craik, D.J.(2000) Biochemistry 39: 15757-15764
- PubMed: 11123900
- DOI: https://doi.org/10.1021/bi000457l
- Primary Citation of Related Structures:
1EWS - PubMed Abstract:
NMR spectroscopy and simulated annealing calculations have been used to determine the three-dimensional structure of RK-1, an antimicrobial peptide from rabbit kidney recently discovered from homology screening based on the distinctive physicochemical properties of the corticostatins/defensins. RK-1 consists of 32 residues, including six cysteines arranged into three disulfide bonds. It exhibits antimicrobial activity against Escherichia coli and activates Ca(2+) channels in vitro. Through its physicochemical similarity, identical cysteine spacing, and linkage to the corticostatins/defensins, it was presumed to be a member of this family. However, RK-1 lacks both a large number of arginines in the primary sequence and a high overall positive charge, which are characteristic of this family of peptides. The three-dimensional solution structure, determined by NMR, consists of a triple-stranded antiparallel beta-sheet and a series of turns and is similar to the known structures of other alpha-defensins. This has enabled the definitive classification of RK-1 as a member of this family of antimicrobial peptides. Ultracentrifuge measurements confirmed that like rabbit neutrophil defensins, RK-1 is monomeric in solution, in contrast to human neutrophil defensins, which are dimeric.
Organizational Affiliation:
Centre for Drug Design and Development, Institute for Molecular Bioscience and Department of Biochemistry, University of Queensland, Brisbane, Queensland 4072, Australia.