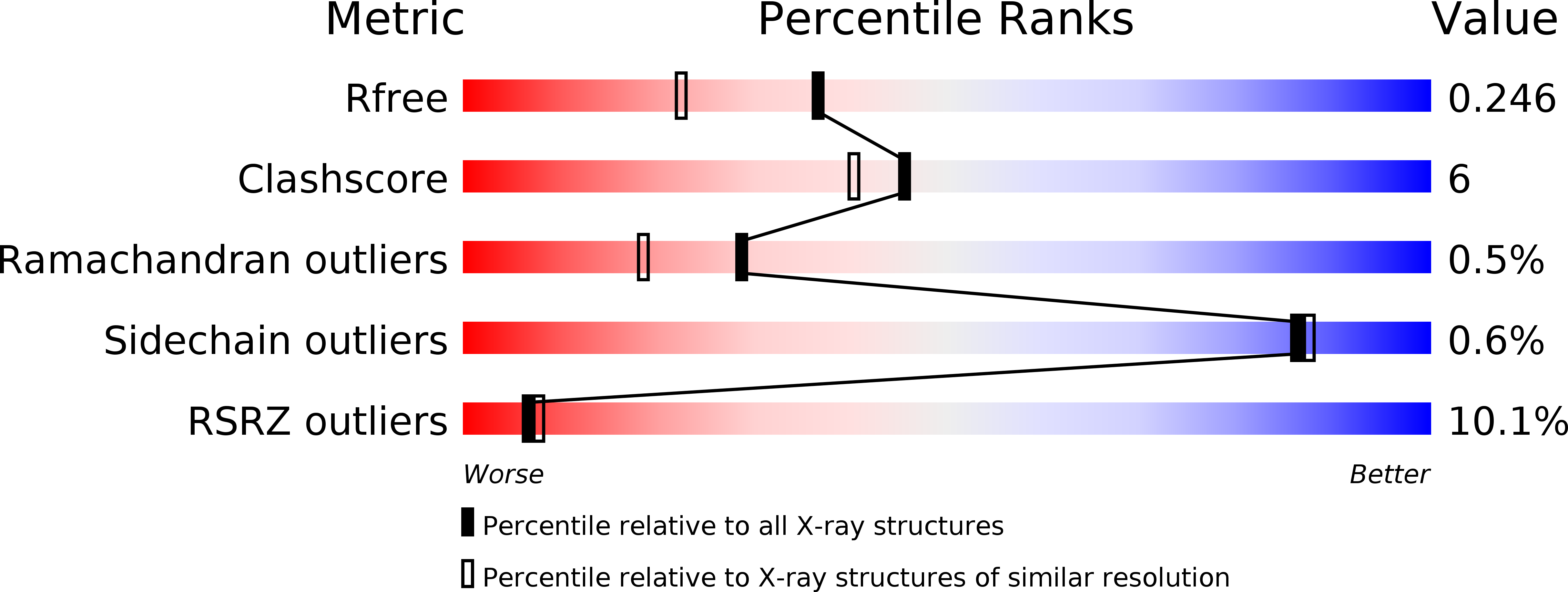
1.9 A resolution crystal structure of the Saccharomyces cerevisiae Ran-binding protein Mog1p.
Stewart, M., Baker, R.P.(2000) J Mol Biol 299: 213-223
- PubMed: 10860733
- DOI: https://doi.org/10.1006/jmbi.2000.3733
- Primary Citation of Related Structures:
1EQ6 - PubMed Abstract:
The 1.9 A resolution X-ray crystal structure of Ran-binding protein Mog1p shows that it has a unique fold based on a six-stranded antiparallel beta-sheet backed on both sides by an extensive alpha-helix. The topology of some elements of Mog1p secondary structure resemble a portion of nuclear transport factor 2 (NTF2), but the hydrophobic cavity and surrounding negatively charged residues that are important in the NTF2-RanGDP interaction are not conserved in Mog1p. In addition to binding RanGTP, Mog1p forms a 1:1 complex with RanGDP and so binds Ran independent of its nucleotide state. Mog1p and NTF2 compete for binding to RanGDP indicating that their binding sites on RanGDP are sufficiently close to prevent both proteins binding simultaneously. Although there may be some overlap between the Mog1p and NTF2 binding sites on RanGDP, these sites are not identical. Sequence analysis of Mog1p homologues from Schizosaccharomyces pombe, human, and Caenorhabditis elegans in the context of the Mog1p crystal structure indicates the presence of a cluster of highly conserved surface residues consistent with an interaction site for Ran.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, England. ms@mrc-lmb.cam.ac.uk