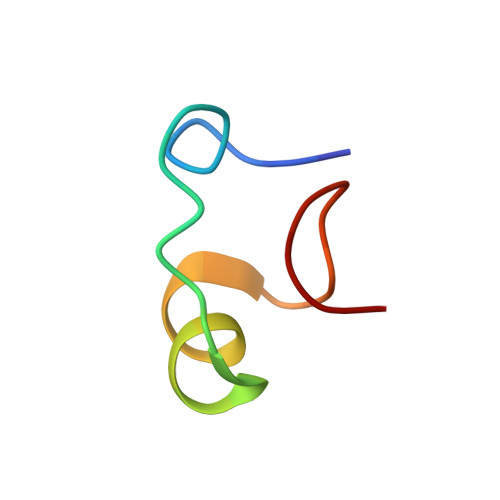
Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Narula, S.S., Brouwer, M., Hua, Y., Armitage, I.M.(1995) Biochemistry 34: 620-631
- PubMed: 7819257
- DOI: https://doi.org/10.1021/bi00002a029
- Primary Citation of Related Structures:
1DMC, 1DMD, 1DME, 1DMF - PubMed Abstract:
Metallothionein is a cysteine-rich metal-binding protein whose biosynthesis is closely regulated by the level of exposure of an organism to zinc, copper, cadmium, and other metal salts. The metallothionein from Callinectes sapidus is known to bind six divalent metal ions in two separate metal-binding clusters. Heteronuclear 1H-113Cd and homonuclear 1H-1H NMR correlation experiments have been used to establish that the two clusters reside in two distinct protein domains. The three-dimensional solution structure of the metallothionein has been determined using the distance and angle constraints derived from these two-dimensional NMR data sets and a distance geometry/simulated annealing protocol. There are no interdomain short distance (< or = 4.5 A) constraints observed in this protein, enabling the calculation of structures for the N-terminal, beta domain and the C-terminal, alpha domain separately. A total of 18 structures were obtained for each domain. The structures are based on a total of 364 experimental NMR restraints consisting of 277 approximate interproton distance restraints, 12 chi 1 and 51 phi angular restraints, and 24 metal-to-cysteine connectivities obtained from 1H-113Cd correlation experiments. The only element of regular secondary structure in either of the two domains is a short segment of helix in the C-terminal alpha domain between Lys42 and Thr48. The folding of the polypeptide backbone chain in each domain, however, gives rise to several type I beta turns. There are no type II beta turns.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut 06520-8066.