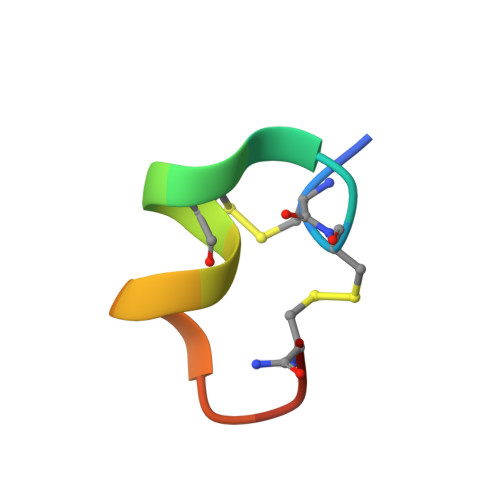
Nuclear magnetic resonance solution conformation of alpha-conotoxin AuIB, an alpha(3)beta(4) subtype-selective neuronal nicotinic acetylcholine receptor antagonist.
Cho, J.H., Mok, K.H., Olivera, B.M., McIntosh, J.M., Park, K.H., Han, K.H.(2000) J Biol Chem 275: 8680-8685
- PubMed: 10722709
- DOI: https://doi.org/10.1074/jbc.275.12.8680
- Primary Citation of Related Structures:
1DG2 - PubMed Abstract:
The neuronal nicotinic acetylcholine receptors constitute a highly diverse group, with subtypes consisting of pentameric combinations of alpha and beta subunits. alpha-Conotoxins are a homologous series of small peptides that antagonize these receptors. We present the three-dimensional solution structure of alpha-conotoxin AuIB, the first 15-residue alpha-conotoxin known to selectively block the alpha(3)beta(4) nicotinic acetylcholine receptor subtype. The pairwise backbone and heavy-atom root mean square deviation for an ensemble of 20 structures are 0.269 and 0.720 A, respectively. The overall fold of alpha-conotoxin AuIB closely resembles that of the alpha4/7 subfamily alpha-conotoxins. However, the absence of Tyr(15), normally present in other alpha4/7 members, results in tight bending of the backbone at the C terminus and effectively renders Asp(14) to assume the spatial location of Tyr(15) present in other neuronal alpha4/7 alpha-conotoxins. Structural comparison of alpha-conotoxin AuIB with the alpha(3)beta(2) subtype-specific alpha-conotoxin MII shows different electrostatic surface charge distributions, which may be important in differential receptor subtype recognition.
Organizational Affiliation:
Protein Engineering Laboratory, Korea Research Institute of Bioscience and Biotechnology, Yusong, Taejon 305-600, Republic of Korea.