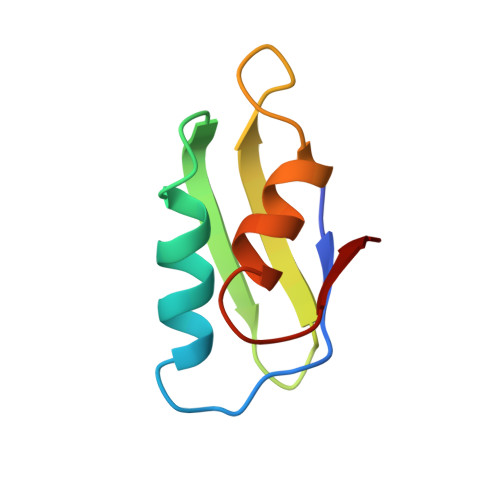
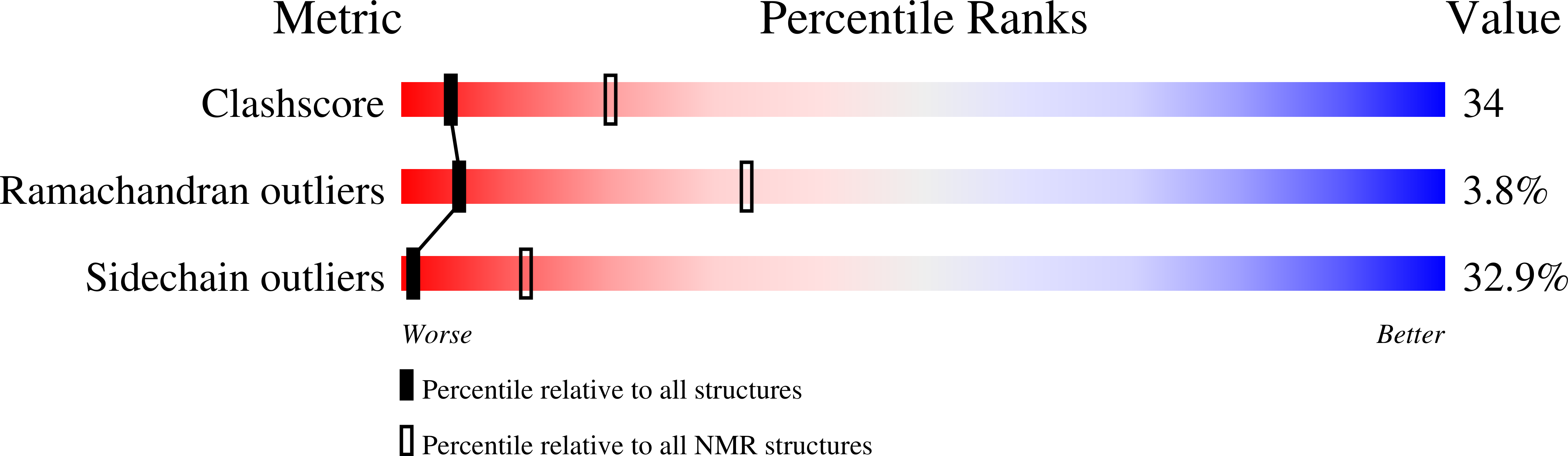NMR structure and metal interactions of the CopZ copper chaperone.
Wimmer, R., Herrmann, T., Solioz, M., Wuthrich, K.(1999) J Biol Chem 274: 22597-22603
- PubMed: 10428839
- DOI: https://doi.org/10.1074/jbc.274.32.22597
- Primary Citation of Related Structures:
1CPZ - PubMed Abstract:
A recently discovered family of proteins that function as copper chaperones route copper to proteins that either require it for their function or are involved in its transport. In Enterococcus hirae the copper chaperone function is performed by the 8-kDa protein CopZ. This paper describes the NMR structure of apo-CopZ, obtained using uniformly (15)N-labeled CopZ overexpressed in Escherichia coli and NMR studies of the impact of Cu(I) binding on the CopZ structure. The protein has a betaalphabetabetaalphabeta fold, where the four beta-strands form an antiparallel twisted beta-sheet, and the two helices are located on the same side of the beta-sheet. A sequence motif GMXCXXC in the loop between the first beta-strand and the first alpha-helix contains the primary ligands, which bind copper(I). Binding of copper(I) caused major structural changes in this molecular region, as manifested by the fact that most NMR signals of the loop and the N-terminal part of the first helix were broadened beyond detection. This effect was strictly localized, because the remainder of the apo-CopZ structure was maintained after addition of Cu(I). NMR relaxation data showed a decreased correlation time of overall molecular tumbling for Cu(I)-CopZ when compared with apo-CopZ, indicating aggregation of Cu(I)-CopZ. The structure of CopZ is the first three-dimensional structure of a cupro-protein for which the metal ion is an exchangeable substrate rather than an integral part of the structure. Implications of the present structural work for the in vivo function of CopZ are discussed, whereby it is of special interest that the distribution of charged residues on the CopZ surface is highly uneven and suggests preferred recognition sites for other proteins that might be involved in copper transfer.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Hönggerberg, CH-8093 Zurich, Switzerland.