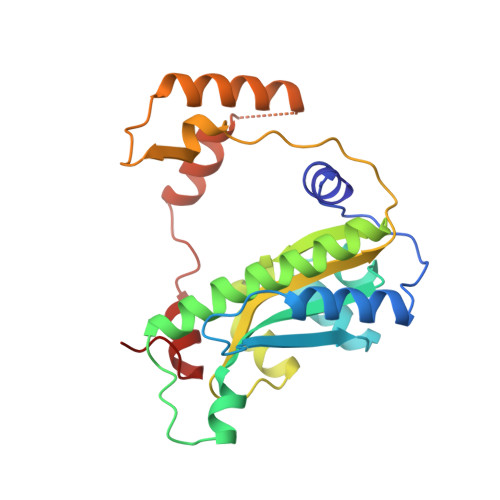
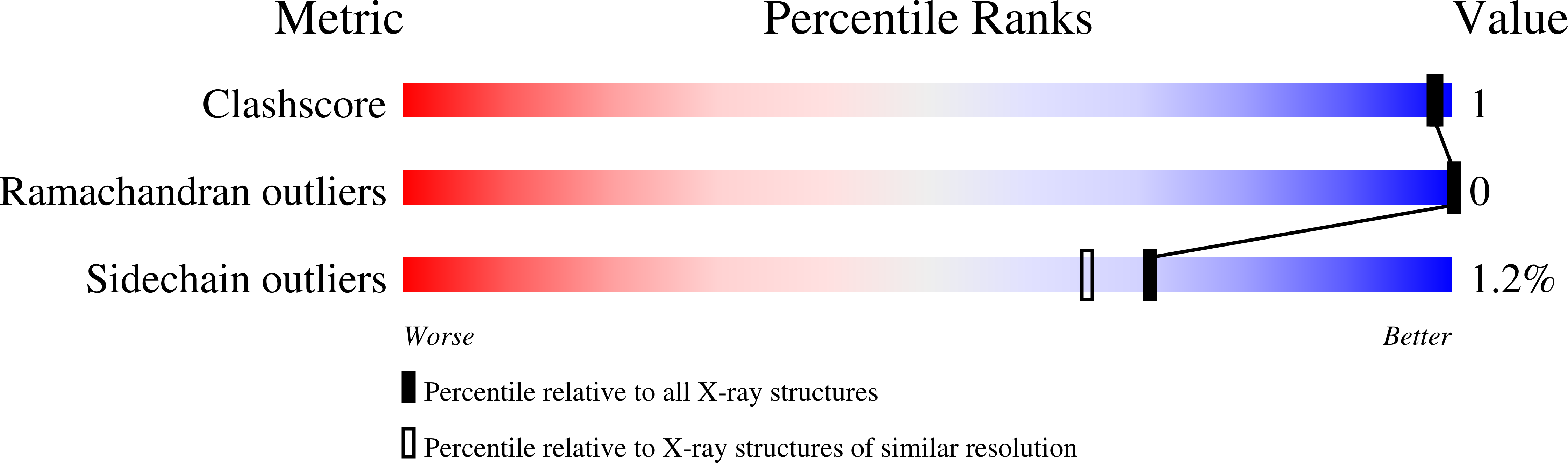Flavin reductase P: structure of a dimeric enzyme that reduces flavin.
Tanner, J.J., Lei, B., Tu, S.C., Krause, K.L.(1996) Biochemistry 35: 13531-13539
- PubMed: 8885832
- DOI: https://doi.org/10.1021/bi961400v
- Primary Citation of Related Structures:
1BKJ - PubMed Abstract:
We report the structure of an NADPH:FMN oxidoreductase (flavin reductase P) that is involved in bioluminescence by providing reduced FMN to luciferase. The 1.8 A crystal structure of flavin reductase P from Vibrio harveyi was solved by multiple isomorphous replacement and reveals that the enzyme is a unique dimer of interlocking subunits, with 9352 A2 of surface area buried in the dimer interface. Each subunit comprises two domains. The first domain consists of a four-stranded antiparallel beta-sheet flanked by helices on either side. The second domain reaches out from one subunit and embraces the other subunit and is responsible for interlocking the two subunits. Our structure explains why flavin reductase P is specific for FMN as cofactor. FMN is recognized and tightly bound by a network of 16 hydrogen bonds, while steric considerations prevent the binding of FAD. A flexible loop containing a Lys and an Arg could account for the NADPH specificity. The structure reveals information about several aspects of the catalytic mechanism. For example, we show that the first step in catalysis, which is hydride transfer from C4 of NADPH to cofactor FMN, involves addition to the re face of the FMN, probably at the N5 position. The limited accessibility of the FMN binding pocket and the extensive FMN-protein hydrogen bond network are consistent with the observed ping-pong bisubstrate--biproduct reaction kinetics. Finally, we propose a model for how flavin reductase P might shuttle electrons between NADPH and luciferase.
Organizational Affiliation:
Department of Biochemical and Biophysical Sciences, University of Houston, Texas 77204-5934, USA.