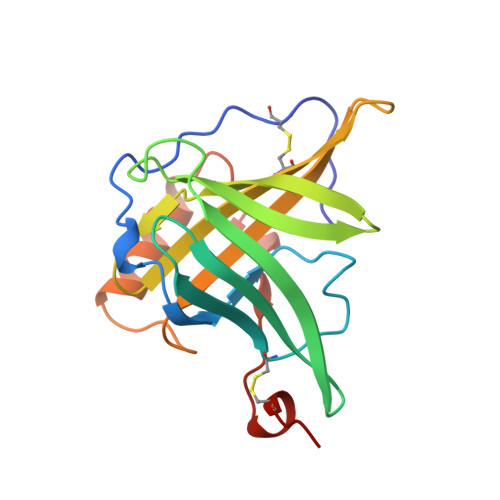
Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution.
Huber, R., Schneider, M., Mayr, I., Muller, R., Deutzmann, R., Suter, F., Zuber, H., Falk, H., Kayser, H.(1987) J Mol Biol 198: 499-513
- PubMed: 3430616
- DOI: https://doi.org/10.1016/0022-2836(87)90296-8
- Primary Citation of Related Structures:
1BBP - PubMed Abstract:
The bilin binding protein (BBP) from the insect Pieris brassicae has been analysed for amino acid sequence, spectral properties and three-dimensional structure. The crystal structure that had been determined by isomorphous replacement has been refined at 2.0 A (1 A = 0.1 nm) resolution to an R-value of 0.20. The asymmetric unit contains four independent subunits of BBP. The co-ordinate differences are 0.25 A, in accord with the estimated error in co-ordinates. The polypeptide chain fold is characterized by an eight-stranded barrel. The connecting loops splay out at the upper end of the barrel and open it, whilst the lower end is closed. The overall shape resembles a calyx. The biliverdin IX gamma chromophore is located in a central cleft at the upper end of the barrel. The bilatriene moiety is in cyclic helical geometry with configuration Z,Z,Z and conformation syn,syn,syn. The geometry is in accord with the spectral properties and permits a correlation between sign of the circular dichroism bands and sense of the bilatriene helices. The fold of BBP is related to retinol binding protein (RBP), as had been recognized in the preliminary analysis, although the amino acid sequences of RBP and BBP show only 10% homology. There are large differences in the loops at the upper end of the barrel, whilst the segments of the centre and the lower end of the barrel superimpose closely. The ligands of BBP and RBP, biliverdin and retinol, respectively, are also similarly located.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Martinsried, BRD.